2025 Vol. 7, No. 46
Foodborne antibiotic-resistant enterococci pose significant risks to One Health and clinical antimicrobial efficacy through food chain transmission. Following the taxonomic reclassification of Enterococcus faecium (E. faecium), comprehensive long-term surveillance data on antibiotic resistance (ABR) patterns and genomic characteristics of E. faecium and Enterococcus lactis (E. lactis) across food animals, environmental sources, and human populations remain limited.
A total of 2,233 samples were collected from multiple nodes along the food chain across 5 Chinese provincial-level administrative divisions (PLADs) during 2015–2019 and 2023–2024. E. faecium (87 isolates) and E. lactis (153 isolates) were identified through whole-genome sequencing and average nucleotide identity analysis. Antimicrobial susceptibility testing, comprehensive genomic content analysis, and pan-genome-wide association studies were performed.
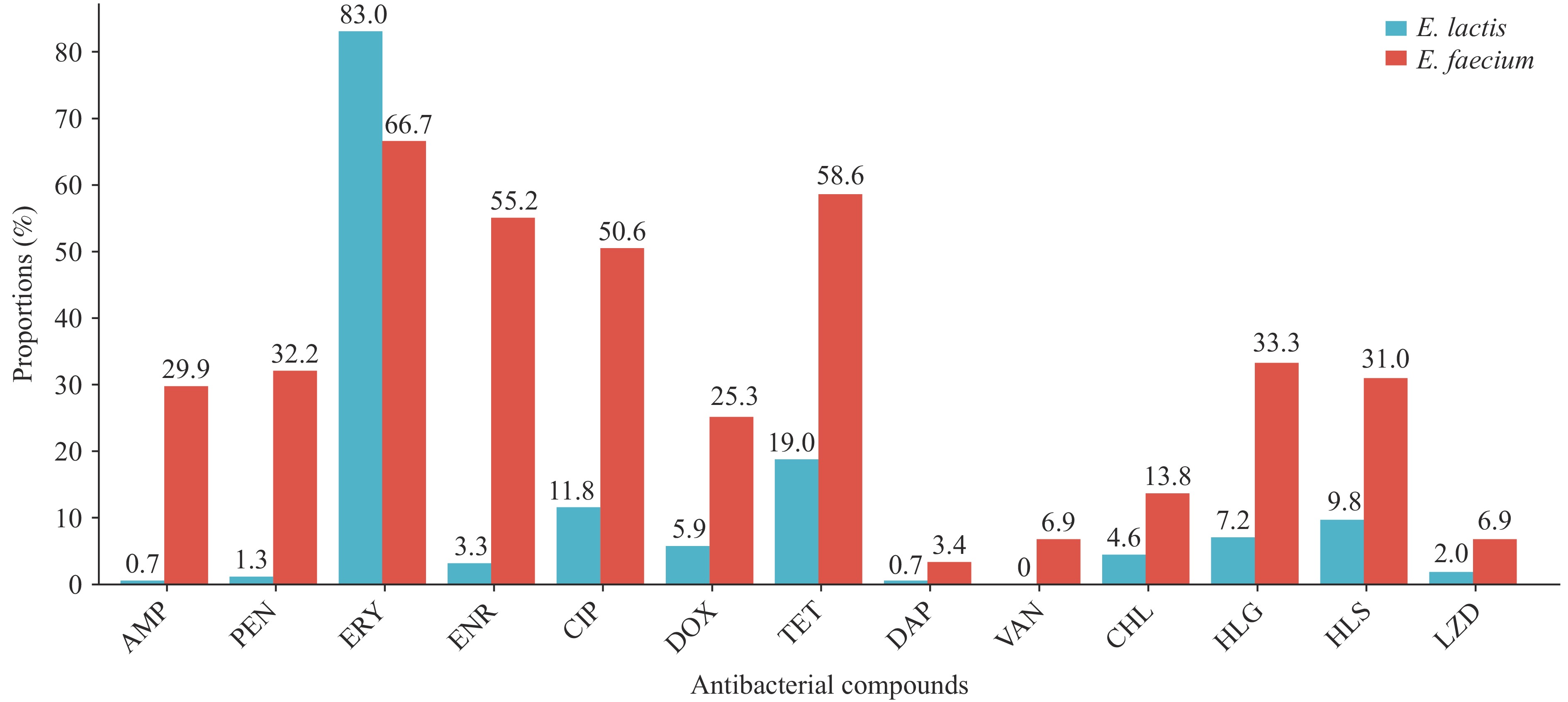
E. faecium demonstrated significantly higher resistance rates to 12 antimicrobials compared with E. lactis (P<0.05). Conversely, E. lactis exhibited a higher resistance rate to erythromycin than E. faecium (P<0.01). The multidrug-resistant (MDR) rate of E. faecium (43/87, 49.4%) substantially exceeded that of E. lactis (16/153, 10.5%) (P<0.001). Genomic analysis revealed that E. faecium harbors significantly more antibiotic resistance genes, mobile genetic elements, and plasmid replicons than E. lactis. No significant interspecies differences were observed in virulence gene profiles associated with adhesion, immune modulation, biofilm formation, and exotoxin production.
E. faecium presents substantially greater ABR risks than E. lactis within the Chinese food chain, necessitating enhanced species-specific surveillance programs. Future interventions should prioritize targeted control strategies tailored to each species to effectively mitigate One Health threats.
Nocardia species are found worldwide in soil rich in organic matter and can cause nocardiosis in humans. Trimethoprim-sulfamethoxazole has long been the first-line treatment for Nocardia infections; however, resistance to this therapy has recently been reported.
Sixty-three clinical Nocardia isolates collected in China were tested against 32 antimicrobial agents using the broth microdilution method. Phylogenetic analysis of the 16S rRNA gene was performed to identify the species.
Three sequences from samples collected in Hainan Province did not match any known Nocardia species, suggesting significant genetic diversity among Nocardia isolates. Nocardia strains generally exhibited high resistance to clarithromycin, clindamycin, and isoniazid. Clinical and reference strains of N. farcinica and N. otitidiscaviarum were susceptible to amikacin and linezolid. Amoxicillin-clavulanate and imipenem were effective against all clinical and reference strains of N. farcinica, whereas gentamicin was effective against all clinical and reference strains of N. otitidiscaviarum.
Linezolid and amikacin were the most consistently active drugs among the analyzed species. Variability of antimicrobial susceptibility was observed among clinical isolates of the same species and between clinical and reference isolates of the same species. Overall, this study highlights the need for better assessment of the burden of nocardiosis in China and for continuous monitoring of antimicrobial resistance among Nocardia isolates.
Asymptomatic carriers of antibiotic-resistant Salmonella constitute a significant yet frequently overlooked public health threat. This study aimed to characterize antimicrobial resistance (AMR) patterns in Salmonella isolated from asymptomatic workers in Yulin, China, over a 12-year period (2013–2024) and to identify the potential influence of natural and socioeconomic factors.
Antimicrobial susceptibility testing was performed against 11 antimicrobial agents. We analyzed temporal trends in AMR rates using the Mann-Kendall test and assessed associations between AMR rates and natural or socioeconomic variables using Spearman’s rank correlation, Principal Component Regression (PCR), and Least Absolute Shrinkage and Selection Operator (LASSO) regression. An Autoregressive Integrated Moving Average (ARIMA) model was employed to forecast future resistance trends.
Resistance to tetracycline (TET) was most prevalent (mean rate: 66.2%). The overall multidrug resistance (MDR) rate was 41.9%, exhibiting a significant increasing trend (P<0.05). Most alarmingly, the tigecycline (TGC) resistance rate surged from 0% to 24.4% by 2024. PCR model analysis revealed that a composite “Socioeconomic and Healthcare Development Index” served as the primary predictor of this increase, explaining 54.9% of the variance in TGC resistance rates. The ARIMA model forecasted a continued upward trajectory for TGC resistance through 2025–2026.
Our findings demonstrate a significant rise in MDR Salmonella among asymptomatic workers in Yulin, establishing them as important reservoirs of antibiotic-resistant Salmonella. The emergence and rapid escalation of TGC resistance is strongly associated with regional socioeconomic and healthcare development. These results underscore the urgent need for integrated surveillance within the One Health framework to effectively address AMR transmission.
The Yellow River serves as a significant conduit for antibiotic resistance transmission from environmental reservoirs to human populations. However, the occurrence and transmission pathways of the clinically relevant extended-spectrum β-lactamase (ESBL) gene blaCTX-M-G9 and ESBL-producing Escherichia coli (E. coli) within the Yellow River remain poorly characterized.
This study reveals the widespread prevalence of blaCTX-M-G9 throughout the Yellow River and its associated water sources in Henan Province, and demonstrates the environmental dissemination and probable animal origin of the dominant E. coli sequence type (ST) 6802 harboring the blaCTX-M-14 genotype.
The findings underscore the critical need to strengthen environmental surveillance and implement robust control measures targeting antibiotic resistance of animal origin. Additionally, we advocate for enhanced public awareness and education initiatives regarding antibiotic resistance to foster broader societal engagement and support for mitigation efforts.
Non-typhoidal Salmonella (NTS) represents a leading cause of foodborne gastroenteritis worldwide. The epidemiological landscape of NTS continues to evolve, with specific serotypes emerging as significant human pathogens through contaminated food products. This evolution occurs particularly within the context of globalized food supply chains and widespread antimicrobial use in agricultural settings.
This study integrated local isolates from Shanghai with global genomic data to reveal distinct international transmission patterns for Salmonella London and Rissen. S. London disseminated through historical, geographically segregated clades, whereas S. Rissen demonstrated recent intercontinental mixing, with Thailand identified as the primary global source. Our analysis identified high-risk plasmids harboring up to 15 resistance genes and demonstrated that Chinese isolates carried the highest antimicrobial resistance burden globally.
The global dissemination of these pathogens is directly linked to international food trade networks. Our findings necessitate a paradigm shift toward integrated global One Health surveillance that bridges human, animal, and food sectors. Implementing harmonized international policies, rigorous trade monitoring, and enhanced antimicrobial stewardship programs is essential to contain the transnational threat posed by resistant foodborne pathogens.



 Subscribe for E-mail Alerts
Subscribe for E-mail Alerts CCDC Weekly RSS Feed
CCDC Weekly RSS Feed