-
Currently, antimicrobial resistance (AMR) is one of the top ten public health threats that not only impacts human public health but also substantially influences animal and environmental health (1). According to a systematic analysis of the past 3 decades (1990‒2021) of global bacterial AMR, it is estimated that 4.71 million deaths associated with bacterial AMR occurred in 2021, including 1.14 million deaths attributable to bacterial AMR (2). In response to this threat, the WHO released the BPPL, which emphasized the prioritization of AMR countermeasures by 2024, highlighting the urgent need to develop novel therapeutics targeting increasingly resistant pathogens (3). This updated list covers 24 antibiotic-resistant pathogens spanning 15 bacterial families, categorizing them into critical, high-, and medium-priority groups, reflecting their global impact. These multidrug-resistant (MDR) pathogens are associated with high morbidity and mortality rates, and therefore, referred to as “superbugs” and are considered a serious threat to global health (4). Based on the bacterial resistance profile and public health impact, the critical antibiotic-resistant ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) had predominantly high scores in the 2024 WHO BPPL list, underscoring their considerable threat to global health (5). Among Gram-negative bacteria, carbapenem-resistant K. pneumoniae (CRKP, top-ranked), third-generation cephalosporin-resistant E. coli, and carbapenem-resistant A. baumannii (CRAB) were among the top three. Carbapenem-resistant P. aeruginosa (CRPA) and Enterobacter spp. were ranked above 60% of the total score. Among the Gram-positive bacteria, vancomycin-resistant E. faecium (VREfm) and methicillin-resistant S. aureus (MRSA) were ranked the highest (5).
-
Given the global threat of AMR, China has implemented a comprehensive range of strategies in alignment with its National Plan on AMR, achieving considerable progress in containment efforts. However, the prevalence of antibiotic-resistant bacteria in clinical settings remains a notable concern (6-7). Based on the CHINET surveillance data of clinical ESKAPE pathogens isolated from approximately 70 hospitals across China over the past decade, although the proportion of ESKAPE strains has remained stable annually (Figure 1A), their absolute numbers showed an increasing trend, in parallel with the overall increase in clinical isolates (Figure 1B). According to the resistance rate data in China, the prevalence of VREfm showed an upward trend from 2019 to 2022 (8). The higher prevalence of MRSA among children has garnered considerable attention despite a decline from 2005 to 2022 (8). As shown in the CHINET data, resistance rates of CRKP, CRAB, and CRPA to carbapenems (imipenem and meropenem) were markedly elevated, exceeding 80% and approaching 100% over the past 4 years (Figure 1C–E). Although the resistance rate of CRKP slightly increased in response to ceftazidime-avibactam, it gained momentum against both colistin and tigecycline, the two main antibiotics used to treat carbapenem-resistant bacteria (Figure 1C). The resistance rate of CRAB remained relatively stable but exhibited a high level of resistance to cefoperazone-sulbactam, a widely employed antimicrobial compound in salvage combination regimens, with a rate exceeding 60% (Figure 1D). The prevalence of CRPA was associated with a notable increase in levofloxacin resistance (from 31.6% in 2021 to 47.9% in 2024) and a growing resistance to piperacillin-tazobactam and ciprofloxacin (Figure 1E).
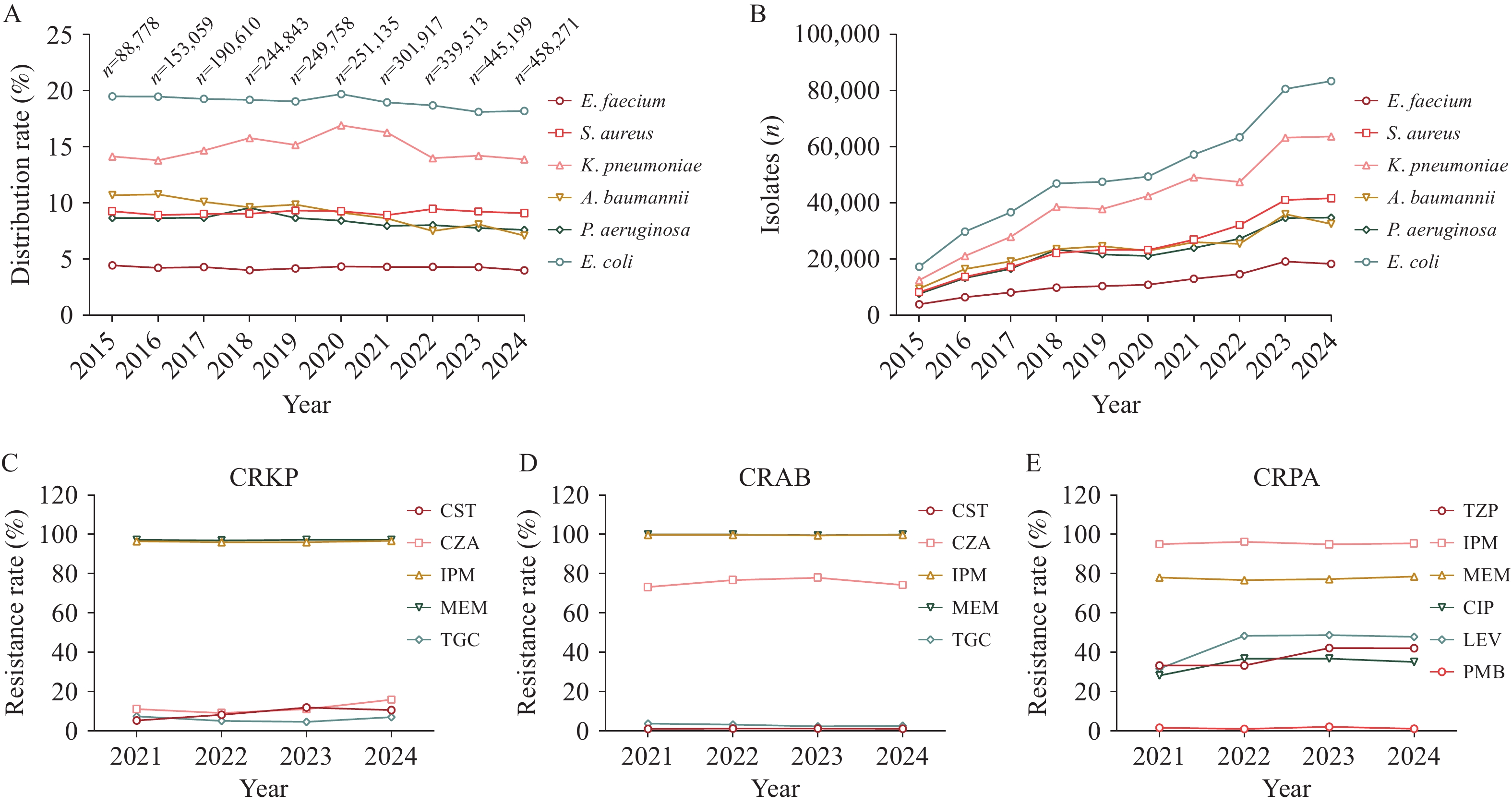 Figure 1.
Figure 1.Trends and antibiotic resistance profile of ESKAPE pathogen clinical isolates in China. (A) Distribution of ESKAPE pathogens from 2015 to 2024; (B) Number of ESKAPE pathogens clinical isolates from 2015 to 2024; (C) Resistance profile of CRKP clinical isolates from 2021 to 2024; (D) Resistance profile of CRAB clinical isolates from 2021 to 2024; (E) Resistance profile of CRPA clinical isolates from 2021 to 2024.
Note: The above data were obtained from CHINET. In panel A, the total number of clinical isolates per year is presented. Abbreviation: CRAB=carbapenem-resistant Acinetobacter baumannii; CST=colistin; CZA=ceftazidime-avibactam; IPM=imipenem; MEM=meropenem; TGC=tigecycline; SCF=cefoperazone-sulbactam; TZP=piperacillin-tazobactam; CIP=ciprofloxacin; LEV=levofloxacin; PMB=polymyxin B; ESKAPE=Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp.The shifting genomic landscape of ESKAPE pathogens poses considerable challenges to public health and clinical medicine owing to the continuous evolution of antibiotic-resistant bacterial clones (9). For example, the prevalence of the VREfm ST80 strain carrying a new type vanA-bearing plasmid is steadily increasing in Guangdong, China (10). Moreover, the genetic diversity of MRSA has been expanding, with the ST59 strain having high potential virulence and emerging as the predominant lineage at 35.6%, surpassing the previously dominant ST239 strain (11–12). The dominant clone of hypervirulent CRKP (Hv-CRKP) ST11 reportedly displays the complexity of MDR, along with genetic heterogeneity in virulence factor profiles, and is considered a health concern owing to its dual threat of high virulence and drug resistance in China (13–14). The widely drug-resistant A. baumannii and P. aeruginosa pose a serious risk to public health, especially in hospital settings, where they can lead to various infections. The widespread clonal ST208 strain, frequently associated with carbapenem resistance genes, is the predominant strain of the extensively drug-resistant A. baumannii (XDRAB), isolated from the intensive care unit (ICU) of a hospital in Jinhua, Zhejiang Province (15). Song et al. reported T3SS-mediated simultaneous secretion of ExoS and ExoU in high-risk P. aeruginosa, which has emerged as a new subset of hypervirulent strains in China (16). Therefore, the complex diversity of emerging ESKAPE highlights the urgent need for careful surveillance of these high-risk strains.
-
To address this challenge, surveillance and mechanistic investigations of bacterial resistance play a crucial role in detecting and tracking the spread of resistant bacteria. Innovations in genome/metagenomic sequencing and analysis technologies could revolutionize AMR surveillance (17). The use of whole-genome sequencing (WGS) of pathogens for AMR surveillance has grown considerably compared with phenotypic surveillance of AMR (18). The construction and interpretation of phylogenetic trees derived from WGS facilitate the identification of transmission events and understanding the dynamics of outbreaks. Using WGS and comparative genome analysis, a multicenter molecular epidemiological survey reported that the high-risk ST11 KL64 CRKP serotype demonstrated substantial expansion potential and survival advantages in China between 2011 and 2021 (19). WGS data can also be used to detect new genomic features, such as AMR genes and virulence genes involved in AMR. A recent multicenter genomics study applied WGS to identify the genomic characteristics and phylogenetic relatedness of CRKP colonization and infection in ICU patients in Anhui Province, China, highlighting the need for coordinated efforts between healthcare facilities and networks to aid CRKP management (20). Moreover, WGS has empowered clinical researchers to identify and monitor AMR while enabling the detection of virulence factors and mobile genetic elements, such as plasmids, which are crucial for understanding the adaptability, pathogenicity, and dissemination of AMR bacteria (18,21). For instance, an additional example of a small blaKPC-2-bearing plasmid essential for the emergence and spread of KPC-2 CRKP from a hospital in Zhejiang, China, was identified upon WGS analysis of a rare CRKP ST437 isolate (22). Furthermore, Liu et al. conducted genetic typing analysis of 90 non-redundant Hv-CRKP isolates from patients and revealed that Hv-CRKP transferability relies on the dominant ST11-K64 clone (23). By analyzing the WGS results of plasmids from 12 representative CRKP isolates, the authors uncovered the clonal spread and clinical evolution of Hv-CRKP within Zhejiang hospitals, involving binary vehicles and 2 fusion plasmid types that facilitate the co-transfer of rmpA hypervirulence and KPC-2 carbapenem resistance (23). Likewise, Huang et al. performed a clinical genomic analysis and reported the diversity and dynamics of blaKPC-2-producing CRPA, providing novel insights into the heterogeneity among CRPA and plasmid-mediated transmission of blaKPC-2 in clinical settings in China (24). Collectively, these studies emphasize the importance of WGS in providing additional insights, thus enhancing epidemiological data and transmission control of AMR pathogens (Figure 2).
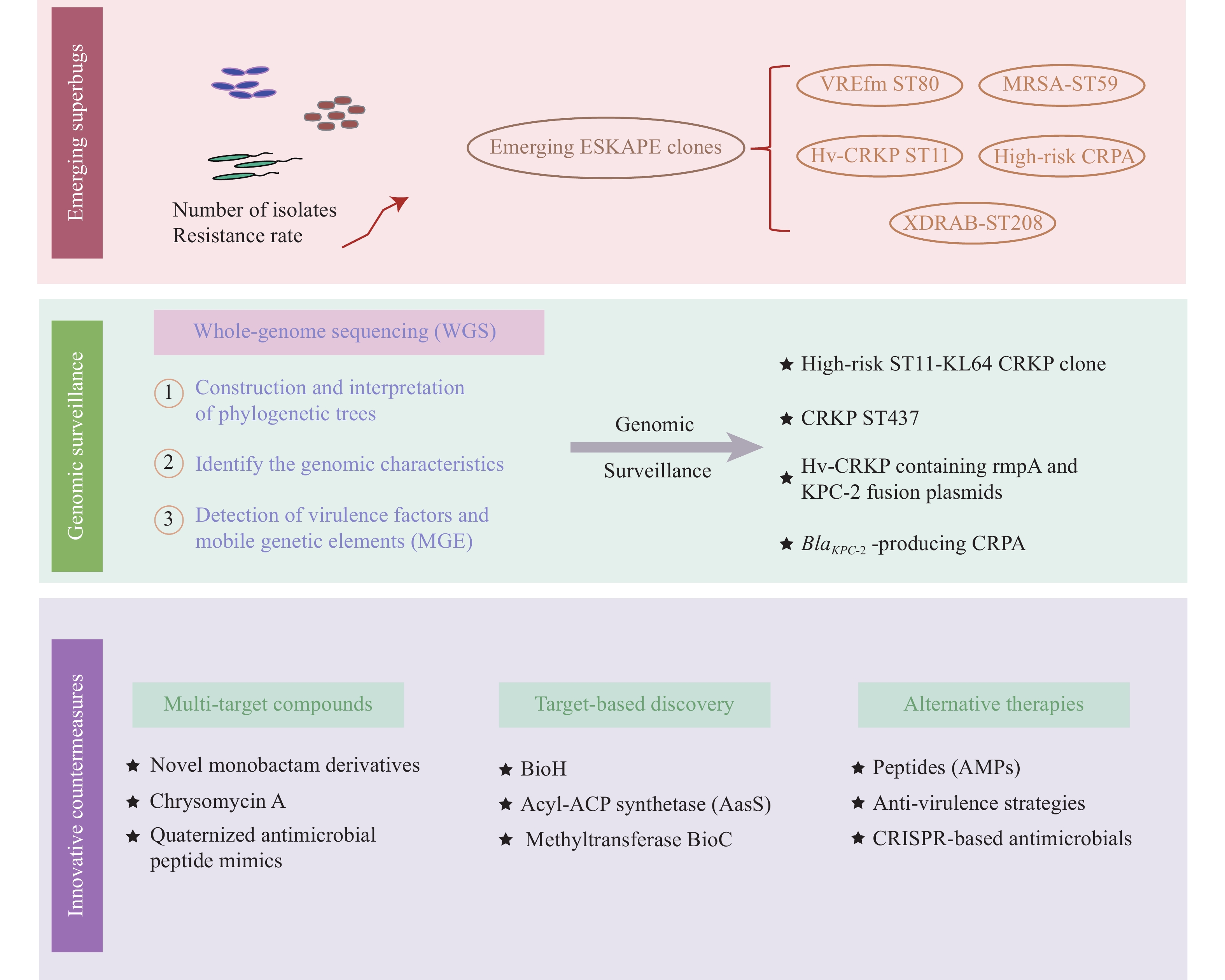 Figure 2.
Figure 2.Structural schematic of the antimicrobial resistance crisis and countermeasures in China.
Abbreviation: AMPs=antimicrobial peptides; CRPA=carbapenem-resistant Pseudomonas aeruginosa; ESKAPE=Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp; MGE=mobile genetic elements; WGS=whole-genome sequencing; CRKP=carbapenem-resistant Klebsiella pneumoniae. -
An effective approach to circumvent existing antibiotic resistance warrants the discovery of new chemical classes, particularly in the fields of natural-product-derived and synthetic small molecules, as well as novel targets and modes of action (25). Compared with single-target drugs, multitarget drugs can simultaneously regulate multiple targets to reduce resistance caused by single-target mutations or expression changes, and have become an effective strategy to combat bacterial resistance (26). A growing body of evidence has shown promise in this regard (Figure 2). For example, Li et al. synthesized a series of novel monobactam derivatives and demonstrated their efficacy against β-lactamase-producing MDR E. coli and K. pneumoniae, with the dual inhibition on PBP3 and class A and C β-lactamases (27). Jia et al. reevaluated chrysomycin A, a novel natural product, which is highly active against MRSA persisters by inhibiting multiple novel targets involved in the biosynthetic pathways of cell wall peptidoglycan and lysine precursors (28). Similarly, multitarget antimicrobial agents quaternized with antimicrobial peptide mimics have been shown to kill MRSA by interacting with lipoteichoic acid and peptidoglycan, causing membrane damage through depolarization, and disrupting cellular redox homeostasis by binding to lactate dehydrogenase (29). Multitarget antibacterial medications are a novel strategy to combat bacterial resistance, and the rational discovery of multitarget drugs may usher in a new golden era for antibiotic discovery.
-
To develop novel antibiotics, target-based strategies must meet the criteria to combat MDR infections. Numerous antibiotic discovery programs have switched to target-based methods to identify substances capable of blocking specific bacterial targets (i.e., essential enzymes or proteins). For instance, the component enzymes of fatty acid or biotin (fatty acid derivatives) synthetic pathways are considered sources of novel antibacterial targets and hold promise for tackling antibiotic-resistant bacteria, such as BioA (30) and BioH (31). Shi et al. discovered that BioH is essential for P. aeruginosa virulence and validated a therapeutic target for reducing CRPA viability, highlighting the potential of inhibiting biotin synthesis following anti-CRPA therapy (32). Huang et al. elucidated the inhibitory mechanism between the acyl adenylate mimic C10-AMS and acyl-ACP synthetase (AasS), which provided a molecular basis for targeting AasS by the C10-AMS inhibitor, thus enabling the re-sensitization of fatty acid synthesis II-targeted antimicrobials (33). Intriguingly, the bacterial methyltransferase BioC, which initiates biotin synthesis, has been identified as a virulence factor of K. pneumoniae, representing an attractive anti-ESKAPE druggable pathway (34).
-
As bacteria become resistant to conventional antibiotics, alternative therapies have been explored in recent years, including antimicrobial peptides (AMPs), anti-virulence strategies, CRISPR-based antimicrobials, and other methods (35). The discovery of novel AMPs is expanding the arsenal of antibacterial drugs. For example, fluorescent 2-phenyl-1 H-phenanthro[9,10-d] imidazole-antimicrobial peptide mimic conjugates have been found to rapidly kill MRSA by disrupting membrane integrity, triggering reactive oxygen species accumulation, causing protein damage, and exhibiting low susceptibility to bacterial resistance (36). Antimicrobial biofilms are expected to efficiently inhibit drug-resistant bacteria. Recently, phenazine-inspired antibiotics were found to be highly active against resistant bacteria, including MRSA, MRSE, and VREfm, by inhibiting biofilm formation (37). A novel gene editing-based antimicrobial strategy was developed using the CRISPR-Cas system to specifically target vital bacterial or resistance genes. The CRISPR-Cas system reportedly targets and eliminates carbapenem-resistant plasmids, thereby restoring antibiotic susceptibility (38). This approach, although still in the experimental stages, offers a potential solution to the challenge of antibiotic-resistant bacteria without the need for novel therapies. Furthermore, the combination of AI and deep learning techniques has the potential to transform drug discovery by accelerating the discovery of novel antibiotic candidates and refining treatment plans based on predictive models of resistance patterns (39).
Although a growing number of antibacterial drugs are under investigation, challenges remain in the development of antimicrobials against drug-resistant pathogens. Currently, approximately 32 new antibacterial compounds are in the clinical trial phase of development, and less than 25% of the drugs in the clinical development pipeline represent a novel class or act through a novel mechanism. Unfortunately, all lack the potential to be effective against major WHO threat pathogens or Gram-negative ESKAPE (40). Hence, developing new antibiotics to combat drug-resistant bacteria remains a challenge.
-
The growing threat of antibiotic resistance warrants a strong and resourceful response based on technological and scientific innovations. Therefore, to preserve antimicrobial efficacy and control resistance transmission, greater efforts should be invested in the discovery and development of novel antimicrobial strategies and in improving comprehensive surveillance systems using next-generation sequencing for source-tracking AMR pathogens. In particular, genomic surveillance involving comprehensive resistance, virulence, and plasmid gene content profiling will enable real-time customization of AMR interventions and address barriers impeding widespread implementation. More importantly, the One Health approach emphasizes the intertwined health of humans, animals, and the environment in disease prevention and control; hence, the control and surveillance of AMR pathogens needs to be considered in all three sectors (1). Coordinated actions across these sectors, such as shared data platforms, joint surveillance programs, and integrated intervention strategies, are pivotal to effectively prevent cross-species transmission and to safeguard public and environmental health in the future (41).
HTML
| Citation: |



 Download:
Download:




