-
Human adenovirus (HAdV) was frequently associated with acute respiratory disease (ARD) outbreaks in closed environment, especially in military camps. In the current study, we investigated the genetic characteristics and epidemiological characteristics of HAdV strains that were responsible for the ARD outbreaks in military camps in China. Among HAdV-related outbreaks that were reported from 2011 to 2014, a total of 3,813 patients were diagnosed from 13,622 camp members, with an overall attack rate of 28.0%. HAdV-B7, HAdV-B14, and HAdV-B55 were determined to be responsible for 3, 2, and 4 outbreaks, respectively. The total attack rate of HAdV-related ARD was 28.0%, ranging from 10.9% to 39.9% among various outbreaks. HAdV-B14 related outbreaks had a higher attack rate than outbreaks of the other two genotypes. Phylogenetic analysis revealed no obvious relationship between outbreak strains and locally circulating strains, indicating the introduction of the strain from a new recruit’s hometown might be responsible for the outbreaks. The findings suggest that vaccine development and administration in military camps must be prioritized. Quarantining among new recruits before entering into the military and the identification of the major responsible genotypes at the current stage is warranted.
Traditionally, there are 51 HAdV serotypes, which are assigned to seven subgroups (A–G) according to biophysical, biochemical, and genetic characteristics. In recent years, new serotypes or subspecies were increasingly recognized by use of phylogenetic analysis, which arised from genome recombination between the hexon gene, fiber, and penton genes. These newly emerged HAdV types resulted in continuous outbreak events on campuses, such as in university campuses or at recruit training centers (1-2), posing severe threats to public health. In China, the ARD outbreak had been constantly reported in the closed community, such as in military camps; however, their etiological agents are rarely determined, in part due to the lack of molecular typing in field investigations. The first known HAdV-associated ARD outbreak in the military was reported from a recruit training camp in 2009, which was determined to be caused by HAdV-B55 infection (3). In this study, we described the HAdV-related ARD outbreaks that had been reported in military camps from 2011–2014, investigating the genetic characteristics of the responsible HAdV strains and their related epidemiological characteristics.
The field investigation and laboratory tests were carried out in accordance with the approved guidelines. Briefly, after the report of an outbreak, nasopharyngeal aspirate (NPA) samples were collected from the index case and from at least 30% of the ARD patients and tested for the commonly seen viral pathogens, i.e., HAdV, influenza virus, respiratory syncytial virus, human rhinovirus, human enterovirus, human coronavirus, and human metapneumovirus. All tests were performed by polymerase chain reaction (PCR) or real-time PCR. HAdV infection from the outbreak was identified by PCR assay using HAdV universal and serotype-specific conventional PCR assays (4-5). To validate the causal relationship between HAdV and the outbreak disease, seroconversion evidence was further determined from paired samples using the neutralizing antibody test. For each outbreak, the responsible HAdV strains were isolated. Briefly, NPA specimens were inoculated into A549 cells incubated at 36 ℃ and observed for cytopathic effects (CPEs) for 21 days. Selected paired samples (acute-phase samples and convalescent-phase samples that were collected over one month apart) were tested for the presence of anti-HAdV antibodies by a colorimetric serum microneutralization assay, using the strains isolated from the outbreak event. All the subjects who were residing in the military camps over the outbreak duration were interviewed using a standard questionnaire to acquire the following information: date of birth, sex, clinical symptoms, date of contact with the patients, and date of onset of symptoms. All data were collected as part of the emergency response to the outbreak events. Data were de-identified for analysis, and the informed consent requirement was waived.
During the period from December 2011 to March 2014, 9 HAdV-related outbreak events of ARD were determined from military camps or recruit training centers, based on both molecular and serological evidences. In each of the outbreaks, other respiratory viruses than HAdV, including influenza A virus or parainfluenza virus, was only occasionally tested as positive, and were not considered as the causative agents of the outbreaks. According to the standard criteria to define ARD, a total of 3,813 patients were diagnosed from 13,622 camp members, with an overall attack rate of 28.0%. The attack rates ranged from 10.9% to 39.9% among various outbreaks (Table 1). Four camps had only male recruits. The other four camps had both male and female recruits, where female patients were also reported, but with a lower attack rate than in males (21.0% vs. 39.4%, P<0.001). The patients were aged from 18 to 24 years. All the HAdV-associated ARD were reported among cold season from December to next March, which coincided with the training period for recruits. The outbreaks were identified to start from 1 to 6 weeks (median 3 weeks) after the initiation of the training activity. The peak outbreak occurred mostly from 11 to 25 days after the disease onset of index case.
Characteristic All participants (N=13,622) ARD patients
(n=3,813)P Gender Male 3,720 (28.35) <0.001 Female 93 (18.56) Age of soldier New recruits 2,722 (31.86) <0.001 Veterans 1,091 (21.48) HAdV genotype HAdV-B7 688 (24.77) <0.001 HAdV-B14 1,016 (37.70) HAdV-B55 2,109 (25.88) Outbreaks areas, province HuBei/2013/HAdV-B7 376 (25.53) <0.001 ShanXi/2013/HAdV-B7 107 (16.54) HuBei/2013/HAdV-B7 205 (31.16) LiaoNing/2012/HAdV-B14 185 (30.32) GanSu/2013/HAdV-B14 831 (39.86) HeBei/2012/HAdV-B55 650 (28.54) ShanXi/2011/HAdV-B55 309 (10.92) TianJin /2012/HAdV-B55 1,150 (37.80) Total 3,813 (27.99) Note: The information on the ninth outbreak of Liaoning /2013/HAdV-B55 was missing. Table 1. Epidemiological characteristics of HAdV-related outbreak events of acute respiratory disease in military camps during December 2011─March 2014.
HAdV-positive results were further confirmed by sequencing of hexon gene, penton, and fiber genes. The genotype was determined by alignment and phylogenetic tree analysis. HAdV-B7, HAdV-B14, and HAdV-B55 were reported to be responsible for 3, 2, and 4 outbreaks, respectively. A higher attack rate was identified from HAdV-B14 (37.7%), significantly higher than that of HAdV-B7 (24.8%) and HAdV-B55 (25.9%) (P<0.001, Table 1). The mean age of the patients was highly comparable among all the outbreak episodes regardless of the HAdV types. The incubation period ranged from 4 to 8 days, showing no difference among outbreaks associated with various genotypes. Among 3,813 ARD patients, 736 (19.3%) developed pneumonia, and the patients with HAdV-B7 (164, 23.8%) and HAdV-B55 (518, 24.6%) had a higher chance of developing pneumonia than those with HAdV-B14 (54, 5.3%; P<0.001).
To investigate the genetic characteristics of the responsible virus, the whole genome sequence of one strain for each outbreak was obtained by deep-sequencing of the HAdV isolates or the raw positive samples. Previously published sequences of HAdV-B7, HAdV-B14, HAdV-B55, and HAdV-B11 were recovered from GenBank and used for the alignment with the current outbreak strains. For each genotype, the phylogenetic trees were respectively constructed based on the HAdV complete genome and 3 segments (penton, hexon, and fiber) by maximum likelihood method using the MEGA 7 software (version 7.0.26, USA) .
For alignment of HAdV-B7 genotype, a total of 109 sequences were obtained from GenBank. Based on the alignment with these available sequences, most of the Chinese strains could be clustered in 5 branches that were distinct from the strains of other countries (Figure 1A, Supplementary Figure S1A, S2A, and S3A). Interestingly all 3-outbreak related HAdV-B7 strains in the current study and the strains from Hubei in the year of 2012 and 2013 formed a separate cluster within the Chinese lineage, irrespective of the geographic location of the outbreak events, possibly suggesting genetic characteristics specific to outbreaks.
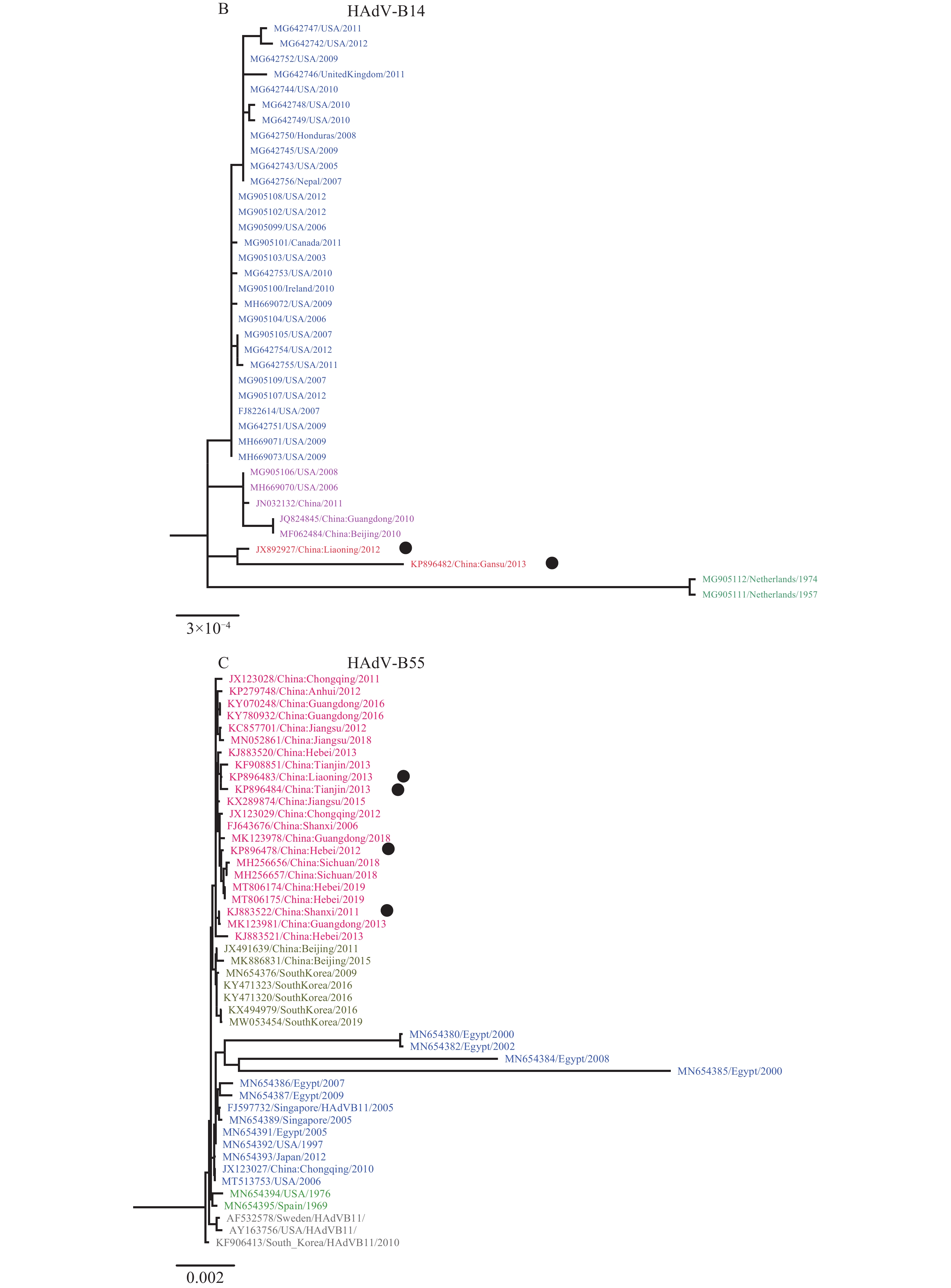 Figure 1.
Figure 1.Phylogenetic tree was constructed based on the full genome sequences using maximum likelihood method with 1,000 bootstraps by MEGA 7.0. (A) HAdV-B7; (B) HAdV-B14; (C) HAdV-B55.
Note: The strains labeled with black dots were obtained from the current study.For HAdV-B14, a total of 50 sequences were recovered from GenBank, including 38 full genome sequences, 5 hexon, and 7 fiber gene sequences. The phylogenetic trees based on full genome sequences demonstrated a clear divergence of the 2 lineages that were derived from the year before 2012 and after 2012 for the strains from China, respectively; while for the hexon gene, the 2 lineages from China became 1 lineage, and for fiber and penton genes, the geographical distribution disappeared (Figure 1B, Supplementary Figures S1B, S2B, and S3B). The strains from China between 2010–2011 were clustered with those from the USA between 2006 and 2008. According to alignment of complete genome sequences, the current outbreak strain (KP896482/China:Gansu/2013) was most closely related to the strain from Liaoning (JX892927/China:Liaoning/2012) (99.9% nucleotide similarity and 99.6% amino acid similarity) while differing from the outbreak strain in the same region that were isolated in 2011 (for hexon gene, JX310315/China:Gansu/2011, 99.8% nucleotide similarity and 98.1% amino acid similarity; for fiber gene, JX310316/China:Gansu/2011, 99.7% nucleotide similarity and 98.1% amino acid similarity). This might reflect that the outbreak event was caused by a strain introduced into the camp by recruits and that the strain did not locally circulate.
For HAdV-B55, a total of 59 sequences were recovered from GenBank, including 47 full genome sequences, 6 hexon, and 6 fiber gene sequences. Based on the full genome sequences, all Chinese strains had high homologies, showing spatial specific distribution patterns and clustering into one branch (Figure 1C). Two strains from Beijing were clustered with strains from the Republic of Korea. While for hexon, fiber, and penton genes, the Chinese strains formed several branches (Supplementary Figures S1C, S2C, and S3C). The strains detected during the recent military outbreaks from 2011 to 2013 were interspersed in the strains isolated from the community strains between 2010 and 2012. This might indicate that the most recent descendant from the early isolates were responsible mainly for the current outbreaks in military camps.
-
In summary, we have determined HAdV-B7, HAdV-B14, and HAdV-B55 to be the most common HAdV genotypes that were responsible for outbreaks in military camps or recruit training centers in China. This is different from the situation in the American military, where HAdV-E4, HAdV-B7, and HAdV-B14 act as the predominant strains sequentially isolated from the outbreaks (6-9). According to prior epidemiological investigation performed in community population in China, HAdV-B3 and HAdV-B7 were most frequently detected among ARD patients, while only in recent years HAdV-B55 emerged with increasing prevalence (10). In the absence of herd immunity against the uncommonly seen HAdV-B14 and newly emerging HAdV-B55 in the local community populations, military recruits were highly susceptible to outbreaks if these 2 types of HAdV were introduced into the susceptible population. An HAdV vaccination program against these three outbreak- related genotypes is urgently needed in this high-risk population.
By comparing various outbreaks, a higher attack rate was identified from HAdV-B14-related outbreaks when compared to those from HAdV-B7 and HAdV-B55, possibly indicating a higher susceptibility and transmissibility of HAdV-B14 in closed settings than the other two types. This conclusion, however, could differ in terms of geographic origin and immunity background of the population. Similar with previous findings (8), younger age and male gender were associated with an increased risk for HAdV infection during outbreak events, irrespective of the HAdV genotypes. Although disease severity varied among genotypes, we observed that infection with HAdV-B7 and HAdV-B55 caused more pneumonia than HAdV-B14 from the outbreak events. The infection source of the HAdV outbreak was preliminarily explored by genetic alignment of the viral strains. For HAdV-B14, no obvious phylogenetic relationship was derived between the HAdV responsible for the outbreak and the locally circulating strain, likely indicating the introduction of the strain from a new recruit’s hometown as the major infectious source. However, whether this hypothesis holds true and how the disease was introduced into the camp are hard to be determined, since the infected individual might still be in the incubation period at the time of entering camp, and thus might be missed even with mandatory quarantine for febrile illness. Quarantine among the new recruits before entering into the military and the identification of the major responsible genotype at the current stage is warranted.
This study was subject to some limitations. The severity of ARD caused by different HAdV genotypes was not evaluated as no individual-level data were collected. The study did not include all the outbreaks of HAdV in closed camps during December 2011–March 2014 in China.
-
No conflicts of interest declared.
HTML
| Citation: |



 Download:
Download:




