-
Hand, foot, and mouth disease (HFMD) is a contagious disease predominantly caused by several enteroviruses and primarily affects infants and children (1). Typically characterized by fever, mouth ulcers, and rash, HFMD symptoms are often mild and generally resolve within 7–10 days. However, some individuals may develop severe symptoms such as fever, a stiff neck, shortness of breath, and worsening rash, leading to potentially life-threatening neurological, respiratory, or circulatory complications, including aseptic meningitis, encephalitis, acute flaccid paralysis, pulmonary hemorrhage, pulmonary edema, and cardiopulmonary failure (2). HFMD was classified as a notifiable disease in May 2008, identifying enterovirus A71 (EV-A71) as the predominant pathogen responsible for severe and fatal cases (3). The widespread administration of an inactivated EV-A71 vaccine in 2016 significantly decreased the incidence of EV-A71-associated HFMD cases (4). Nevertheless, the diversity of pathogens leading to severe HFMD has evolved recently due to the absence of cross-protection among different enterovirus serotypes (5). Coxsackievirus A6 (CVA6) has emerged as the primary pathogen in severe HFMD cases across various regions in China. For instance, in 2013, the Third People’s Hospital of Shenzhen reported that all eight patients with severe HFMD tested positive for CVA6, all developing meningitis, with two also suffering myocardial damage (6). Similarly, in 2017, the Guangdong Women and Children’s Hospital found that among 55 patients with CVA6-associated severe HFMD, 29 (52.7%) developed aseptic meningitis, and six (10.9%) also experienced pulmonary edema (7).
Enteroviruses possess a single-stranded, positive-sense RNA genome that encodes both structural and nonstructural viral proteins. After entering a cell, the genomic RNA translates into a polyprotein divided into three regions: P1, P2, and P3. This polyprotein is then segmented into individual proteins by viral proteases. Specifically, the P1 region comprises four structural proteins (VP1–VP4) (8). Variations in the nucleotide sequences of the VP1 region are utilized for molecular typing of enteroviruses (9). Studies focusing on the molecular typing of CVA6 based on these sequences have identified the D3 sub-genotype as the predominant strain globally since 2008, with the D3a sub-genotype being the most widespread in China (5).
Based on the national laboratory surveillance network for HFMD pathogens established in the Chinese mainland in 2008, a total of 74 CVA6 strains were obtained from severe HFMD cases between 2012 and 2023. Analysis of their VP1 sequences allowed for the inference of the population's historical dynamics and the evolutionary characteristics of CVA6. This study aims to provide insights into the surveillance of severe HFMD cases.
-
According to the HFMD Treatment Guidelines (2010 and 2018 editions), severe HFMD cases were defined by the following clinical signs: persistent high fever (>39 °C), neurological symptoms (depression and abnormal movement), atypical respiratory symptoms (abnormalities in respiratory rate), and circulatory dysfunction (abnormal heart rate and prolonged capillary refill time). Cases that met these criteria were included in the study, which aimed to enhance public health surveillance and informed decision-making without involving human experimentation. The study received approval from the Second Ethics Review Committee of the National Institute for Viral Disease Control and Prevention at the Chinese Center for Disease Control and Prevention.
-
According to surveillance guidelines, the local CDC collected samples (e.g., stool and throat swabs) from severe HFMD cases and transported them to designated laboratories for EVs screening using Real-time RT-PCR. All CVA6-positive samples were subsequently forwarded to the provincial CDC for virus isolation following the standard protocol (Polio Laboratory Manual, 4th ed, https://iris.who.int/handle/10665/68762). The National Polio Laboratory at the National Institute of Viral Disease Prevention and Control in China was tasked with genotype identification.
-
Nucleic acid extraction from cell cultures was carried out using the Tianlong nucleic acid extraction kit (Ex-DNA/RNA Virus (CDC)/T327, Xi’an Tianlong Technology Co., Ltd., China) and the GeneRotex 96 nucleic acid extractor (Xi’an Tianlong Technology Co., Ltd., China). The VP1 coding region was amplified through reverse transcription polymerase chain reaction (RT-PCR) utilizing the PrimeScript One Step RT-PCR kit version 2 (RR057A, TaKaRa, China). Specific primers for amplification included CVA6-2339Y (5’–3’: CCTTCTGAGGCCAACATCAT) and CVA6-3461Z (5’–3’: ATACCAAGTTGGCCCAGTCA). Sequencing was conducted using an ABI 3130 genetic analyzer (Applied Biosystems, Foster City, CA, USA), and data analysis was performed using Sequencher software (version 5.4.6, Ann Arbor, USA) to determine the VP1 coding sequence of enteroviruses. Molecular typing of sequences was completed via the enterovirus typing tool available at www.rivm.nl/mpf/enterovirus/typingtool.
-
The alignment of sequences was performed using MAFFT software (version 7.490) (10), while the best nucleotide substitution model was identified using ModelGenerator (version 0.85). RAxML-NG (version 0.9.0) was utilized to construct the maximum likelihood phylogenetic tree (11). TempEst (version 1.5) facilitated the analysis of the temporal structure of the sequences (12). The optimal molecular clock model and tree prior were determined through Path Sampling/Stepping-stone techniques (13). Bayesian phylogenetic analysis was executed using BEAST (version 1.8.4) (14), with the analysis outputs reviewed in Trace software (version 1.7.1). The Bayesian maximum clade credibility (MCC) tree was constructed using TreeAnnotator (version 1.8.4) and visualized in FigTree software (version 1.4) (http://tree.bio.ed.ac.uk/software/). A single haplotype was defined in DnaSP6 software (version 6.12.03) (15) when nucleotide sequences were identical, aiding in the understanding of nucleotide mutations throughout viral evolution. The median-joining haplotype network was built using PopART software (version 1.7).
-
This study analyzed 74 CVA6 strains isolated from severe HFMD cases, which were submitted by provincial HFMD surveillance laboratories between 2012 and 2023. Of the 74 CVA6-associated severe HFMD cases, 48 were in males and 26 in females. The patients had a median age of 2.0 years (mean age 2.05 years, range 6 months to 6.5 years). The age distribution was as follows: 25 cases in children under 1 year old, 26 cases in those aged 1–5 years, and three cases in children older than 5 years (Table 1).
Patient number Gender Age (years) Region Year Haplotype Sub-genotype NMDC number HFMD1 Female 1 Northwest China 2018 Hap_41 D3a NMDCN00038LF HFMD2 Male 1 Northwest China 2017 Hap_37 D3a NMDCN00038L8 HFMD3 Female 2 Northwest China 2015 Hap_42 D3a NMDCN00038LI HFMD4 Male 4 Northwest China 2015 Hap_43 D3a NMDCN00038LJ HFMD5 Female 3 Northwest China 2015 Hap_29 D3a NMDCN00038JS HFMD6 Female 1 South China 2021 Hap_48 D3a NMDCN00038K8 HFMD7 Male 2 South China 2023 Hap_56 D3a NMDCN00038LH HFMD8 Female 2 South China 2023 Hap_57 D3a NMDCN00038LN HFMD9 Male 2 South China 2023 Hap_58 D3a NMDCN00038LQ HFMD10 Male 4 South China 2020 Hap_47 D3a NMDCN00038K5 HFMD11 Male 2 Southwest China 2022 Hap_62 D3a NMDCN00038LU HFMD12 Male 4 Southwest China 2022 Hap_63 D3a NMDCN00038LV HFMD13 Male 4 Southwest China 2022 Hap_63 D3a NMDCN00038M0 HFMD14 Male 3 Southwest China 2022 Hap_64 D3a NMDCN00038M1 HFMD15 Female 1 Southwest China 2022 Hap_60 D3a NMDCN00038LR HFMD16 Male 2 Southwest China 2022 Hap_61 D3a NMDCN00038LS HFMD17 Female 2 Southwest China 2020 Hap_59 D3a NMDCN00038LC HFMD18 Female 5 South China 2018 Hap_54 D3a NMDCN00038KS HFMD19 Female <1 South China 2018 Hap_55 D3a NMDCN00038KT HFMD20 Female 2 South China 2018 Hap_55 D3a NMDCN00038KU HFMD21 Male 1 South China 2018 Hap_53 D3a NMDCN00038KR HFMD22 Female 2 South China 2018 Hap_51 D3a NMDCN00038KM HFMD23 Female 1 South China 2018 Hap_52 D3a NMDCN00038KN HFMD24 Male 5 South China 2018 Hap_49 D3a NMDCN00038KK HFMD25 Male 2 South China 2018 Hap_50 D3a NMDCN00038KL HFMD26 Male <1 North China 2017 Hap_25 D3a NMDCN00038KO HFMD27 Female <1 North China 2017 Hap_26 D3a NMDCN00038KQ HFMD28 Male 2 North China 2017 Hap_24 D3a NMDCN00038KJ HFMD29 Male 1 North China 2015 Hap_23 D3a NMDCN00038K2 HFMD30 Male 1 Central China 2019 Hap_14 D3a NMDCN00038L3 HFMD31 Male /* Central China 2020 Hap_6 D3a NMDCN00038K3 HFMD32 Female − Central China 2020 Hap_12 D3a NMDCN00038L0 HFMD33 Male − Central China 2020 Hap_9 D3a NMDCN00038KB HFMD34 Male − Central China 2020 Hap_10 D3a NMDCN00038KC HFMD35 Male − Central China 2020 Hap_11 D3a NMDCN00038KP HFMD36 Female − Central China 2020 Hap_3 D3a NMDCN00038JU HFMD37 Male − Central China 2020 Hap_4 D3a NMDCN00038JV HFMD38 Male − Central China 2020 Hap_5 D3a NMDCN00038K1 HFMD39 Male − Central China 2020 Hap_7 D3a NMDCN00038K4 HFMD40 Male − Central China 2020 Hap_7 D3a NMDCN00038K6 HFMD41 Male − Central China 2020 Hap_8 D3a NMDCN00038K9 HFMD42 Male − Central China 2020 Hap_1 D3a NMDCN00038JR HFMD43 Male − Central China 2020 Hap_15 D3a NMDCN00038L4 HFMD44 Male − Central China 2020 Hap_17 D3a NMDCN00038L9 HFMD45 Male − Central China 2020 Hap_18 D3a NMDCN00038LD HFMD46 Male − Central China 2020 Hap_19 D3a NMDCN00038LG HFMD47 Female − Central China 2020 Hap_20 D3a NMDCN00038LK HFMD48 Female − Central China 2020 Hap_20 D3a NMDCN00038LO HFMD49 Male − Central China 2020 Hap_13 D3a NMDCN00038L1 HFMD50 Female 6 Central China 2019 Hap_16 D3a NMDCN00038L6 HFMD51 Male 4 Central China 2014 Hap_2 D3a NMDCN00038JT HFMD52 Male 4 East China 2017 Hap_21 D3a NMDCN00038L7 HFMD53 Female 1 East China 2017 Hap_22 D3a NMDCN00038LT HFMD54 Female 3 Northwest China 2020 Hap_32 D3a NMDCN00038KE HFMD55 Male 4 Northwest China 2020 Hap_33 D3a NMDCN00038KF HFMD56 Male <1 Northwest China 2013 Hap_35 D3a NMDCN00038KH HFMD57 Male 1 Northwest China 2013 Hap_45 D3a NMDCN00038LM HFMD58 Female 3 Northwest China 2013 Hap_39 D3a NMDCN00038LB HFMD59 Female 1 Northwest China 2013 Hap_36 D3a NMDCN00038KI HFMD60 Male 1 Northwest China 2013 Hap_32 D3a NMDCN00038KD HFMD61 Male 1 Northwest China 2013 Hap_31 D3a NMDCN00038K7 HFMD62 Female 1 Northwest China 2013 Hap_30 D3a NMDCN00038K0 HFMD63 Female 2 Northwest China 2013 Hap_46 D3a NMDCN00038LP HFMD64 Male 1 Northwest China 2013 Hap_44 D3a NMDCN00038LL HFMD65 Male 1 Northwest China 2013 Hap_40 D3a NMDCN00038LE HFMD66 Male 2 Northwest China 2013 Hap_38 D3a NMDCN00038LA HFMD67 Male 1 Northwest China 2013 Hap_37 D3a NMDCN00038L5 HFMD68 Male 2 Northwest China 2013 Hap_34 D3a NMDCN00038KG HFMD69 Male 1 Northwest China 2013 /† D2 NMDCN00038KA HFMD70 Male 1 Northwest China 2013 − D2 NMDCN00038L2 HFMD71 Male 1 Northwest China 2013 Hap_27 D3a NMDCN00038JP HFMD72 Female 1 Northwest China 2014 Hap_36 D3a NMDCN00038KV HFMD73 Female 2 Northwest China 2014 Hap_28 D3a NMDCN00038JQ HFMD74 Male − Southwest China 2021 Hap_65 D3a NMDCN00038M2 Note: Northwest China includes Shaanxi, Gansu, and Qinghai provinces; and Ningxia Hui Autonomous Region and Xinjiang Uygur Autonomous Region. South China includes Guangdong and Hainan provinces; Guangxi Zhuang Autonomous Region; and Hong Kong SAR and Macao SAR. Southwest China includes Sichuan, Guizhou, and Yunnan provinces; Chongqing Municipality; and Xizang Autonomous Region. North China includes Hebei and Shanxi provinces; Beijing and Tianjin Municipality; and Inner Mongolia Autonomous Region. Central China includes Henan, Hubei, and Hunan provinces. East China includes Jiangsu, Zhejiang, Anhui, Fujian, Jiangxi, and Shandong provinces; Shanghai Municipality; and Taiwan, China.
Abbreviation: HFMD=hand, foot and mouth disease; SAR=Special Administrative Region.
* Missing age information for this case;
† No haplotype analysis of D2.Table 1. Summary of information on 74 cases of CVA6-associated severe hand, foot, and mouth disease in China, 2012–2023.
CVA6 isolates identified in 97.3% (72/74) of cases were predominantly of the D3a sub-genotype, while the D2 sub-genotype was solely found in Northwest China (Figure 1). The cases were primarily located in Central and Western China, with the most cases reported in Northwest China (25 cases), followed by Central China (22 cases), South China (13 cases), Southwest China (8 cases), North China (4 cases), and East China (2 cases). Incidences before 2015 were predominantly in Northwest China, whereas occurrences post-2015 were mainly in Central, South, and Southwest China (Table 1).
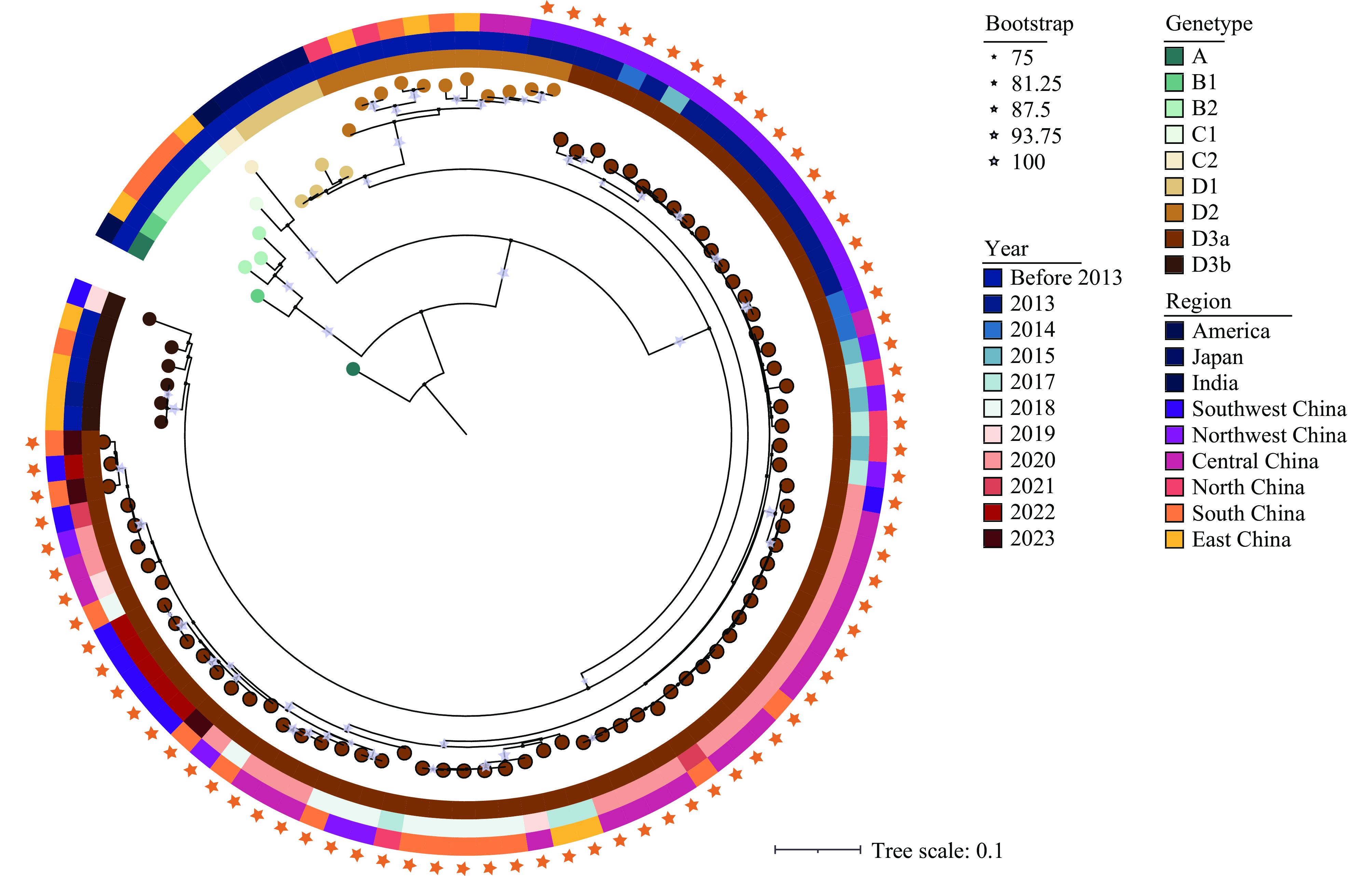 Figure 1.
Figure 1.Molecular typing of 74 CVA6 strains isolated from severe HFMD in China, 2012–2023. The ML tree, constructed using the VP1 coding region, was utilized to determine the genotype of the CVA6 strains isolated in this study.
Abbreviation: CVA6=Coxsackievirus A6; HFMD=hand, foot, and mouth disease; ML=maximum likelihood. -
The MCC tree analysis revealed that CVA6 sequences from the same geographical region displayed high similarity and often clustered together. Regarding the temporal distribution of CVA6 in severe HFMD cases, sampling in Northwest China was primarily conducted before 2015. In contrast, post-2015 samples predominantly came from patients with severe HFMD in Central, South, and Southwest China. The evolutionary distances between post-2015 sequences and those isolated earlier in Northwest China significantly increased. Specifically, the average genetic distance was 0.027 for sequences before 2015 and increased to 0.051 for sequences post-2015 compared to earlier sequences (Figure 2A). Within the same region, the average genetic distance among sequences was 0.032, whereas it was 0.044 between different regions (Figure 2B). Furthermore, Bayesian skyline plot analysis indicated that the D3a sub-genotype of CVA6, isolated from 72 cases, experienced three population expansions during its evolutionary history, specifically in 2012–2013, 2013–2014, and 2019–2020. Notably, there were significant increases in the population size during the periods 2012–2013 and 2019–2020 (Figure 2C).
 Figure 2.
Figure 2.Inferred historical population dynamics of 74 CVA6 strains isolated from severe HFMD cases in China, 2012–2023. (A) Variation in nucleotide sequences of CVA6 from different regions. (B) Variation in nucleotide sequences of CVA6 across different years. (C) MCC tree and Bayesian skyline plot of the VP1 region of CVA6 depicting the 95% confidence intervals of the HPD analysis with light blue shading. Sequences from various regions are differentiated by color. (D) In the median-joining network, the circle size correlates with haplotype frequency, while unsampled haplotypes are shown as small black solid circles. Each connecting line indicates a mutational step between haplotypes.
Abbreviation: CVA6=Coxsackievirus A6; HFMD=Hand, foot, and mouth disease; MCC=Maximum clade credibility; HPD=Highest posterior density.Analysis of the 72 CVA6 D3a VP1 sequences identified 258 variable sites within the 915 bp fragment, encompassing 65 haplotypes. In 2013, there were 14 haplotypes, with 41 additional haplotypes emerging over the subsequent decade. Of the 65 haplotypes, 58 (89.2%, 58/65) comprised solely a single sequence. These haplotypes were distributed across various regions, forming several large clusters of regionally originated haplotypes in the haplotype network plot and the MCC tree. This clustering indicates that haplotypes from different regions diverged through multiple base substitutions. The analyses also imply the existence of further undetected samples, as evidenced in Figure 2D and Table 1.
-
Unlike mild HFMD, severe cases can lead to neurological, respiratory, or circulatory complications, and treatment delays may result in further deterioration. EV-A71 has been identified as the predominant pathogen responsible for severe HFMD. However, following the introduction of the inactivated EV-A71 vaccine in 2016, the spectrum of pathogens causing HFMD has shifted, with other enteroviruses, particularly CVA6, emerging as the primary causative agents (5).
Seventy-four cases of severe HFMD associated with CVA6 were reported in children under the age of 5 years. The immature immune systems of this age group may heighten their susceptibility, leading to severe complications and potentially fatal outcomes if not promptly diagnosed and treated (2). Consequently, the development and administration of vaccines targeting CVA6 are crucial to prevent severe HFMD in susceptible children.
The CVA6 genotype, particularly the dominant D3a sub-genotype in China, has been isolated primarily, with only two instances of the D2 strain identified in 2013. This underscores the necessity for ongoing robust surveillance of severe HFMD specifically targeting the CVA6 D3a sub-genotype. In this study, a total of 74 CVA6 strains were collected from severe HFMD cases between 2012 and 2023, predominantly in Central China, Northwest China, and South China. It is important to note that the limited number of samples and the considerable variability in surveillance quality across provinces may introduce bias in the analysis, potentially skewing the true prevalence of CVA6-associated severe HFMD.
D3a has been the predominant sub-genotype of CVA6 in China since 2012. Population dynamic analyses reveal that the virus isolated from severe HFMD cases exhibited three major expansions, reflecting increased diversity within the CVA6 population post-2012. Haplotype reconstruction from 74 sequences identified 65 unique haplotypes, underscoring the extensive diversity among CVA6 strains associated with severe HFMD across different regions. Notably, no shared haplotypes were found between regions, suggesting the existence of undetected haplotypes and potentially unmonitored severe HFMD cases associated with CVA6. Current diagnostic criteria for severe HFMD, which include mild clinical symptoms with neurological or circulatory complications (2), may lead to misdiagnoses, such as enteroviral meningitis, thus contributing to the underreporting of severe HFMD cases. Given these findings, enhancing surveillance and improving clinician training on the identification and reporting of severe HFMD is essential to better assess and manage the disease burden.
-
No conflicts of interest.
-
The staff of the local Centers for Disease Control and Prevention in Gansu, Guangdong, Guizhou, Hainan, Hebei, Henan, Hunan, Jiangsu, Shandong, Shaanxi, Sichuan, Yunnan, Zhejiang, and Chongqing PLADs for their efforts in collecting the clinical samples.
HTML
Case Inclusion Criteria
Sample Collection and Viral Isolation
Molecular Typing of Enteroviruses
Inference of Population Historical Dynamics
Demographic Characteristics of HFMD Cases with CVA6 Infections
Inference of Population Historical Dynamics
| Citation: |



 Download:
Download:




