-
On May 9, 2022, 3 local residents in Guang’an City, Sichuan Province, tested positive for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and were diagnosed as asymptomatic infections with coronavirus disease 2019 (COVID-19) in a local outbreak, and as of May 14, a total of 20 confirmed cases and 398 asymptomatic infections were reported.
Whole genome sequencing was performed directly on the nasal and throat swab specimens from the initial three asymptomatic infected individuals using the Illumina MiSeq platform (Illumina, San Diego, CA, USA). The complete genome sequences of 3 SARS-CoV-2 strains, named SC1215, SC1216, and SC1217, were obtained. The sequence of SC1217 has been deposited in the GISAID database (under the accession number EPI_ISL_12725020). Phylogenetic analysis (maximum likelihood method) revealed that the 3 local strains clustered together to form a monophyletic clade belonging to the Omicron BA.2.2 sublineage (Figure 1) (1–2). The BA.2.2 sublineage was derived from the Omicron BA.2 lineage (3) and may have originated in Hong Kong Special Administrative Region (SAR) with predominant transmission to the mainland of China, Japan, Australia, and the United Kingdom (2).
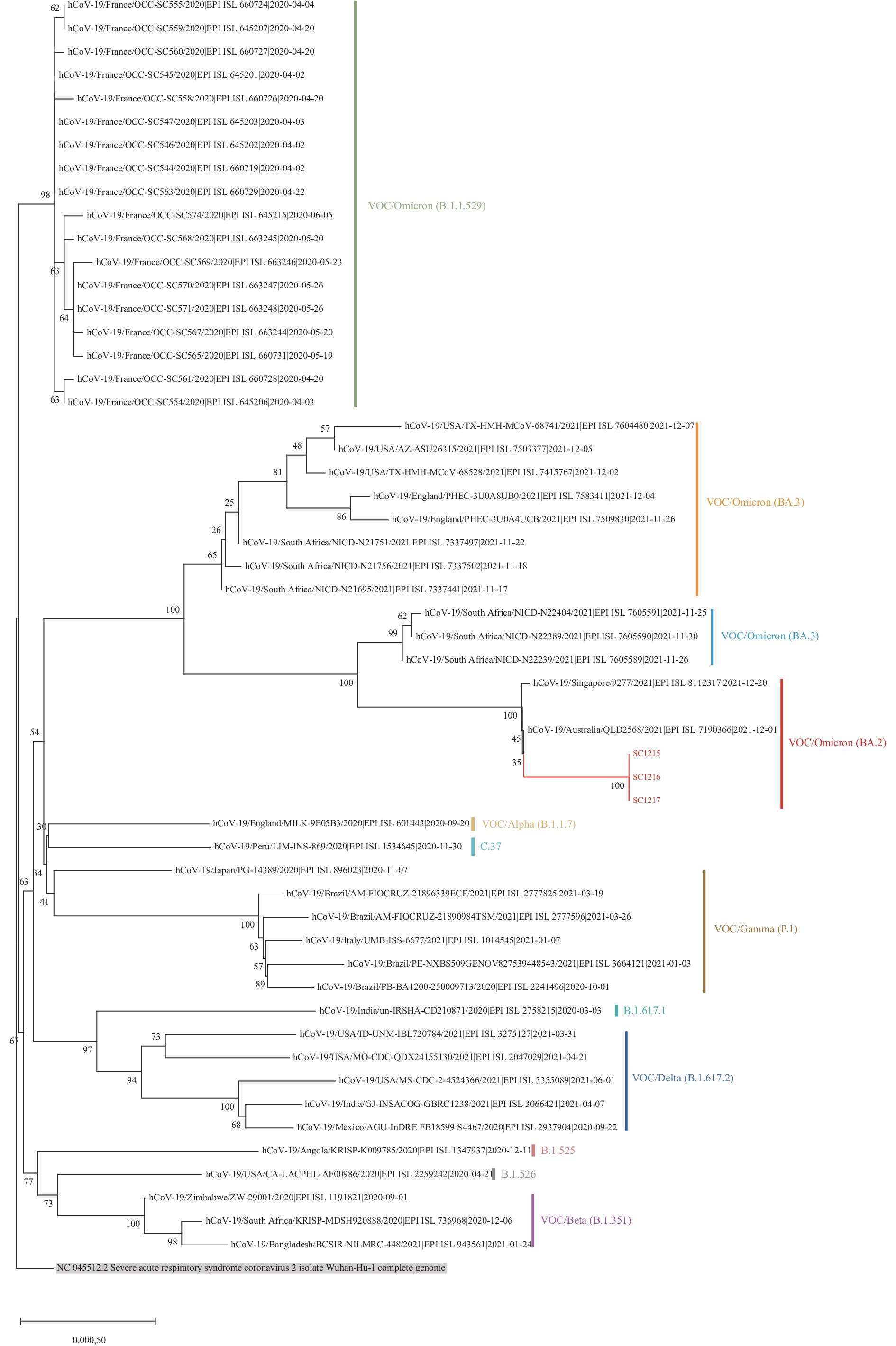 Figure 1.
Figure 1.The maximum likelihood phylogeny based on the full-length genome sequences of SARS-CoV-2.
Note: The Sichuan 454-nt deletion variants are highlighted in red. The Wuhan reference strain is shaded in gray.
Abbreviation: VOC=variant of concern; SARS-CoV-2=severe acute respiratory syndrome coronavirus 2.
Genomic analysis revealed a 454 nucleotide (nt) deletion in the SARS-COV-2 genome at nucleotide positions 27,689–28,142, and this result was further validated by Sanger sequencing (Figure 2). Sequencing of 41 clinical specimens from the local outbreak in Guang’an by May 14 had also detected the 454-nt deletion. Further analysis revealed that the 454-nt deletion resulted in a 71-nt deletion at the 3’ end of ORF7a gene, a complete deletion of ORF7b gene (132-nt) and the intergenic region of ORF7b/8 (6-nt) gene, and a 249-nt deletion at the 5’ end of ORF8 gene.
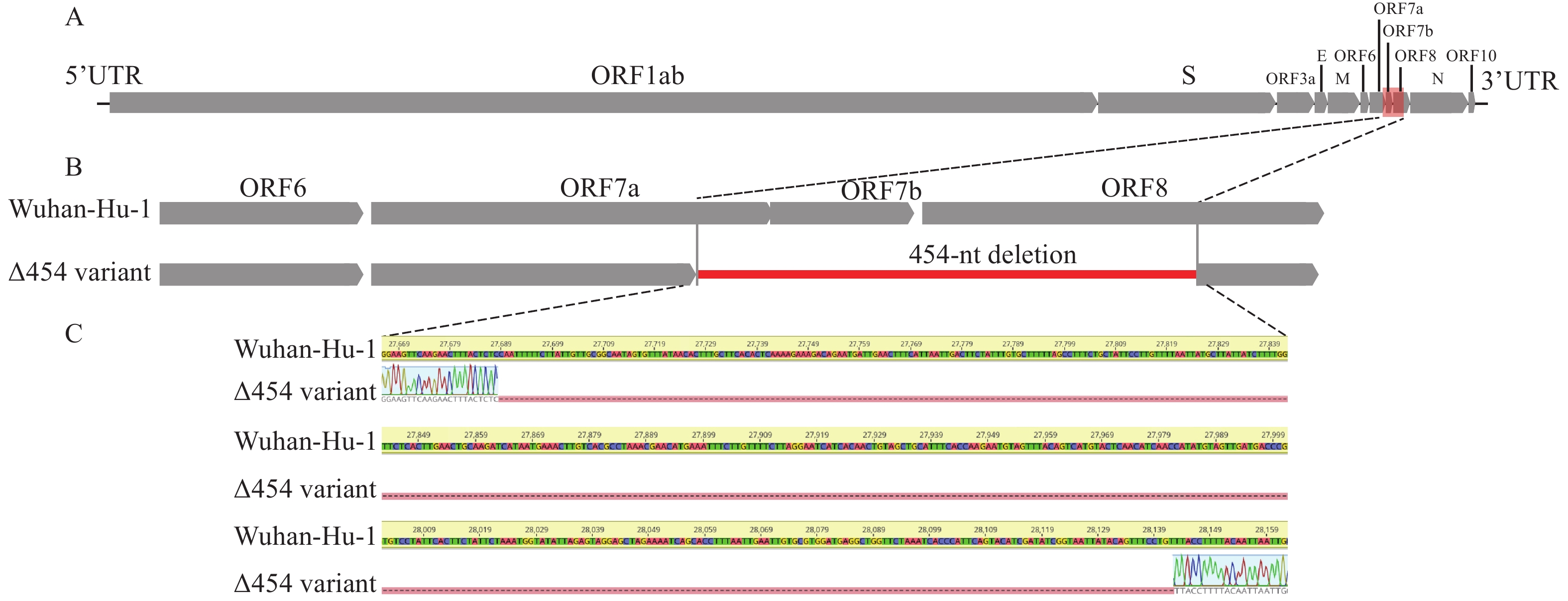 Figure 2.
Figure 2.Genomic organization of human SARS-CoV-2 and the location of the 454-nt deletion(Δ454). (A) Schematic diagram of the genomes of SARS-CoV-2 isolates Wuhan-Hu-1 (Genbank: NC_045512, GISAID: EPI_ISL_402125). (B) Magnification of genomic region (red box in panel A) showing the 454-nt deletion spanning ORF7a, ORF7b, and ORF8 (indicated by the red line). (C) Sanger sequencing results flanking the 454-nt deletion of the SARS-CoV-2 Δ454 variant.
Abbreviation: SARS-CoV-2=severe acute respiratory syndrome coronavirus 2.As of May 12, 2022, 2,787 sequences in the GISAID database were Omicron BA.2.2 sublineage, but no such long 454-nt deletion were found. ORF7a, ORF7b, and ORF8, which encode accessory proteins of SARS-CoV-2, are important for the interaction of SARS-CoV-2 with the host. Previous studies have shown that ORF7a protein binds to human monocytes, decreases antigen presentation ability, and induces dramatic expression of pro-inflammatory cytokines (4-5). Deletion of ORF8 may lead to a milder infection and a more efficient immune response to SARS-CoV-2 (6). Further experiments are needed on whether and how this 454-nt deletion affects the infectivity and pathogenicity of the virus, and epidemiological and genomic surveillance is needed to monitor its potential further transmission.
HTML
| Citation: |



 Download:
Download:




