-
It remains inconclusive whether short-term ozone exposure can cause an inflammatory response and oxidative damage in the circulatory system. We conducted a longitudinal panel study of 43 non-smoking college students with four rounds of visits in Shanghai Municipality, China, from May to October 2016. Personal real-time ozone exposure was monitored for 3 days per visit, followed by blood sample collection. We measured 10 inflammatory biomarkers, 2 oxidative stress biomarkers, and 2 indicators of mitochondrial oxidative damage in the blood. Linear mixed-effect (LME) models were used to analyze their associations with ozone in R software (V 3.5.2, R Foundation for Statistical Computing, Vienna, Austria) and a P-value <0.05 was considered statistically significant. During the study period, mean ozone exposure levels ranged from 18.0 ppb to 22.7 ppb at different lag periods. Each 10-ppb increase in ozone exposure was greatly associated with a 4.86% increase in tumor necrosis factor alpha (TNF-α), 3.14% in soluble intercellular adhesionmolecule-1 (sICAM-1), and 0.89% in malondialdehyde (MDA), as well as a decrease of 0.23 in the average methylation (%5mC) of the mitochondria displacement loop (D-loop) region. Our results suggest that even short-term exposure to low-level ozone may lead to inflammation, lipid peroxidation, and mitochondrial oxidative damage.
For each visit, personal ozone exposure was monitored in real time for 3 days during daytime (from 8∶00 to 18∶00) using Personal Ozone Monitors (POMs, 2B Technologies, US). In addition, we used HOBO data loggers (Onset Computer Corporation, Pocasset, MA, US) to monitor temperature and relative humidity at the individual level. For each individual, we collected blood samples immediately after ozone monitoring. Serum levels of 10 inflammatory biomarkers were measured using the MILLIPLEX MAP Human Cytokine/Chemokine Kit (Millipore Corp., Billerica, MA, US), including interleukin-6 (IL-6), interleukin-8 (IL-8), interleukin-10 (IL-10), interleukin-17 alpha (IL-17α), TNF-α, monocyte chemoattractant protein-1 (MCP-1), granulocyte macrophage colony stimulating factor (GM-CSF), sICAM-1, soluble vascular cell adhesion molecule-1 (sVCAM-1), and vascular endothelial growth factor (VEGF). Overall, 2 markers of oxidative stress, including 8-isoprostaglandin F2 alpha (8-iso-PGF2α) and MDA, were measured in serum using enzyme linked immunosorbent assay (Oxford Medical, UK). We also measured the relative mitochondrial DNA copy number (RMtDNAcn) and the methylation of the mitochondria D-loop region in blood to indicate mitochondrial oxidative damage, according to previous methods (1). The study was approved by the Institutional Review Board of the School of Public Health, Fudan University (No. 2014-07-0523). All participants signed informed consent at enrollment.
We applied LME models to analyze the associations between personal ozone exposure and these biomarkers, with each participant’s identification number incorporated as random intercepts. Data on 10 inflammatory biomarkers, 2 oxidative stress biomarkers, and RMtDNAcn were naturally log-transformed before statistical analyses. To capture the lagged effects of ozone, we considered exposure windows of 0–2 hours, 3–5 hours, 6–8 hours, 0–8 hours, 1 day, and 2 days preceding the blood sample collection. We also included: 1) individual characteristics, including age, sex, body mass index (BMI); 2) two natural cubic smooth functions of temperature and relative humidity with 3 degrees of freedom (dfs) for both; and 3) a natural cubic smooth function of the day within the study period with 3 dfs. We further performed a sensitivity analysis by adjusting for fine particulate matter, sulfur dioxide, nitrogen dioxide, and carbon monoxide, separately.
Three asthmatic patients were excluded, leaving 40 participants (30 females and 10 males) for the current analysis. Their mean age was 24 years, and their mean BMI was 21 kg/m2. As shown in Supplementary Table S1, the average personal ozone exposure at different lags ranged from 17.8±20.5 ppb to 22.7±17.4 ppb, which was far below the current ambient air quality standard in China (i.e., 8-hour maximum: 160 μg/m3, equivalent to 75 ppb). The mean levels of 14 biomarkers varied appreciably, and detailed descriptive data can be found in Table 1.
Biomarker Mean SD Min Median Max IQR Inflammation IL-6 (pg/mL) 1.40 1.35 0.41 0.91 10.01 0.90 IL-8 (pg/mL) 9.51 7.18 2.74 6.93 33.22 8.67 IL-10 (pg/mL) 2.05 4.00 0.21 0.70 24.53 0.90 IL-17α (pg/mL) 9.33 16.50 0.82 2.96 122.67 6.86 TNF-α (pg/mL) 7.27 4.21 1.70 6.65 24.57 4.33 MCP-1 (pg/mL) 392.32 156.15 160.24 361.94 1,005.00 181.06 GM-CSF (pg/mL) 6.76 10.36 0.72 1.87 50.88 5.32 sICAM-1 (ng/mL) 133.18 69.43 25.79 112.08 440.74 45.44 sVCAM-1 (ng/mL) 519.97 131.90 34.15 504.76 907.56 178..27 VEGF (pg/mL) 205.32 333.76 7.35 88.17 1,737.00 154.40 Oxidative stress 8-iso-PGF2α (ng/mL) 0.56 2.46 0.01 0.16 19.80 0.20 MDA (μmol/L) 29.26 2.45 16.81 29.35 35.83 2.76 Mitochondrial oxidative damage RMtDNAcn 1.00 2.05 0.00 0.62 20.71 0.42 Methylation (%5mC)* 2.55 2.15 0.00 2.47 8.62 3.99 Abbreviations: SD=standard deviation; Min=minimum; Max=maximum; IQR=interquartile range; pg=picogram; ng=nanogram; IL-6=interleukin-6; IL-8=interleukin-8; IL-10=interleukin-10; IL-17α=interleukin-17 alpha; TNF-α=tumor necrosis factor alpha; MCP-1=monocyte chemoattractant protein-1; GM-CSF=granulocyte macrophage colony stimulating factor; sICAM-1=soluble intercellular adhesionmolecule-1; sVCAM-1=soluble vascular cell adhesion molecule-1; VEGF=vascular endothelial growth factor; 8-iso-PGF2α=8-isoprostaglandin F2 alpha; MDA=malonaldehyde; RMtDNAcn=relative mitochondrial DNA copy number.
* Methylation levels of the D-loop region in mitochondrial DNA.Table 1. Summary statistics on biomarkers of inflammation, oxidative stress, and mitochondrial oxidative damage in 40 college students in Shanghai Municipality, China, May–October 2016.
Lag Mean SD P5 P25 Median P75 P95 0–2 h 22.2 14.2 4.5 12.3 19.8 28.1 53.1 3–5 h 22.7 17.4 4.5 11.2 18.2 30.3 65.6 6–8 h 18.0 20.5 4.5 7.6 13.3 21.3 53.6 0–8 h 21.0 14.7 4.5 11.5 17.4 26.1 54.1 1 d 19.4 11.2 5.2 10.9 18.4 25.9 40.7 2 d 19.6 11.5 6.0 10.7 17.3 25.7 40.8 Abbreviations: SD=standard deviation; P5=5th percentile; P25=25th percentile; P75=75th percentile; P95=95th percentile. Table S1. Descriptive statistics on personal ozone exposure during different periods preceding the blood sample collection in Shanghai Municipality, China, May–October 2016.
Item Ozone + PM2.5 Ozone + SO2 Ozone + NO2 Ozone + CO TNF−α 4.62 (0.82, 8.56) 4.07 (0.24, 8.05) 4.29 (0.32, 8.42) 4.75 (0.76, 8.90) sICAM−1 3.21 (0.30, 6.20) 3.32 (0.35, 6.38) 3.06 (0.05, 6.16) 2.60 (−0.41, 5.69) sVCAM−1 2.05 (−0.72, 4.91) 2.25 (−0.05, 4.59) 2.11 (−0.23, 4.50) 1.94 (−0.46, 4.40) VEGF 10.57 (−0.10, 22.38) 10.89 (0.24, 22.63) 9.87 (−0.74, 21.60) 10.12 (−0.53, 21.90) MDA 0.90 (0.09, 1.72) 0.88 (0.10, 1.66) 0.88 (0.09, 1.67) 0.87 (0.08, 1.65) RMtDNAcn 30.51 (−3.77, 76.99) 31.45 (0.07, 72.66) 27.82 (−2.70, 67.92) 30.86 (−0.41, 71.94) Methylation† −0.24 (−0.48, 0.01) −0.22 (−0.43, −0.01) −0.24 (−0.45, −0.02) −0.25 (−0.47, −0.04) Abbreviations: TNF−α=tumor necrosis factor alpha; sICAM−1=soluble intercellular adhesionmolecule−1; sVCAM−1=soluble vascular cell adhesion molecule−1; VEGF=vascular endothelial growth factor; MDA=malonaldehyde; RMtDNAcn=relative mitochondrial DNA copy number; PM2.5=ambient particulate matter with aerodynamic diameter less than 2.5 μm; SO2=sulfur dioxide; NO2=nitrogen dioxide; CO=carbon monoxide.
* Lag 3–5 hours for MDA and lag 0–2 hours for other biomarkers.
† Methylation levels of the D−loop region in mitochondrial DNA.Table S2. Percentage changes in biomarkers with a 10-ppb increase in personal ozone exposure* in 40 college students in Shanghai Municipality, China, May–October 2016, with adjustments of other air pollutants on the day of the blood sample collection.
Among the 10 inflammatory biomarkers, we observed significant increases in TNF-α, sICAM-1, sVCAM-1, and VEGF with ozone exposure (Figure 1). However, we did not find any significant associations of ozone with IL-6, IL-8, IL-10, IL-17α, MCP-1, GM-CSF (Supplementary Figure S1). We also found that ozone exposure showed a positive and statistically significant association with MDA, while the association with 8-iso-PGF2α was insignificant (Figure 2). In addition, we observed a significant increase in RMtDNAcn but a significant decrease in the D-loop methylation with ozone exposure (Figure 2).
 Figure 2.
Figure 2.Percentage changes in 8-iso-PGF2α (A), MDA (B), and RMtDNAcn (C), and absolute change in mitochondria D-loop methylation levels (D) associated with a 10-ppb increase in personal ozone exposure at different lag periods in 40 college students in Shanghai Municipality, China, May–October 2016.
Note: X-axis denotes to the different lag periods; Y-axis denotes the corresponding percentage/absolute change in biomarkers. Abbreviations: 8-iso-PGF2α=8-isoprostaglandin F2 alpha; MDA=malonaldehyde; RMtDNAcn=relative mitochondrial DNA copy number.The lag patterns of these significant associations were similar, except for the association with MDA. These effects mostly occurred within 2 hours of exposure, then were gradually attenuated over longer lags, and lost statistical significance at a lag of 2 days. At lag 0–2 hours, a 10-ppb increase in ozone concentrations was associated with the following increases: 4.86% [95% confidence interval (CI): 1.39%–8.45%] in TNF-α; 3.14% (95% CI: 0.87%–5.46%) in sICAM-1; 2.23% (95% CI: 0.09%–4.40%) in sVCAM-1; 10.26% (95% CI: 0.65%–20.79%) in VEGF; and 30.51% (95% CI: 0.31%–69.81%) in RMtDNAcn. A decrease of 0.23 (95% CI: 0.01%–0.45%) was found in the D-loop methylation (%5mC). The significant association with MDA was observed at lag 3–5 hours only, with a 0.89% (95% CI: 0.14%–1.65%) increase in MDA per 10-ppb increase in ozone exposure at this lag. After adjusting for other air pollutants, the associations of ozone with TNF-α, sICAM-1, MDA, and the D-loop methylation were almost unchanged, while the associations with sVCAM-1, VEGF, and RMtDNAcn were unstable (Supplementary Table S2).
-
In this longitudinal panel study, we found that short-term exposure to low concentrations of ozone may lead to increased biomarkers of inflammation and oxidative stress, including TNF-α, sICAM-1, and MDA. We also observed reduced methylation of the mitochondria D-loop region with ozone exposure.
It remains inconclusive whether ozone exposure could induce an inflammatory response in the circulatory system, although extensive evidence suggested that short-term ozone exposure was associated with respiratory inflammation. Consistent with previous studies (2-3), we found ozone exposure was associated with increased sICAM-1 in this study. An increased circulating level of sICAM-1 was relevant to endothelial injury due to inflammation and may be an independent risk factor for atherosclerosis and cardiovascular disease (4-5). In addition, we found ozone exposure was associated with elevated TNF-α, a marker of systemic inflammation. However, we did not find any significant associations with other common markers of systemic inflammation (i.e., interleukins, MCP-1, and GM-CSF). Previous studies have also showed mixed results on the inflammatory effects of ozone (6-7), and further studies are needed to confirm our findings.
Mitochondrial DNA is sensitive to reactive oxygen species as it lacks protective histones and DNA repair mechanisms. Mitochondrial DNA methylation is an indicator of mitochondrial oxidative damage (1). To the best of our knowledge, this is the first study to find an association between ozone exposure and hypomethylation of the mitochondria D-loop region. This finding suggests that ozone exposure may trigger mitochondrial damage and dysfunction, thereby disrupting cellular homeostasis and leading to metabolic alterations (8). We also observed an increase in RMtDNAcn, another indicator of mitochondrial oxidative damage, with ozone exposure. However, the observed increase became statistically insignificant when adjusting for other air pollutants. Notably, these pollutants were measured by fixed-site monitoring rather than personal monitoring, and therefore one should be cautious in interpreting the results of the two-pollutant models. In addition, we found that ozone exposure was associated with increased MDA, which is one of the final products of polyunsaturated fatty acids peroxidation and plays a role in the development of cardiovascular disease (9). A previous study found increased 8-iso-PGF after exposure to high levels of ozone (10), but the results of this study did not report such an association at low concentrations.
Our study was subject to at least three limitations. First, the statistical power may be restricted due to the small sample size. As a result, the confidence intervals for the effect estimates in this study were wide. Second, the participants were only college students, which may limit the generalizability of our findings to other populations and settings. Third, all health outcomes were measured at the same time, restricting the ability to evaluate the causality between ozone exposure and blood biomarkers.
In conclusion, our study found that even short-term exposure to low concentrations of ozone was associated with inflammation, lipid peroxidation, and mitochondrial oxidative damage. Our results suggest that the current air quality standards for ozone need to be further tightened in China.
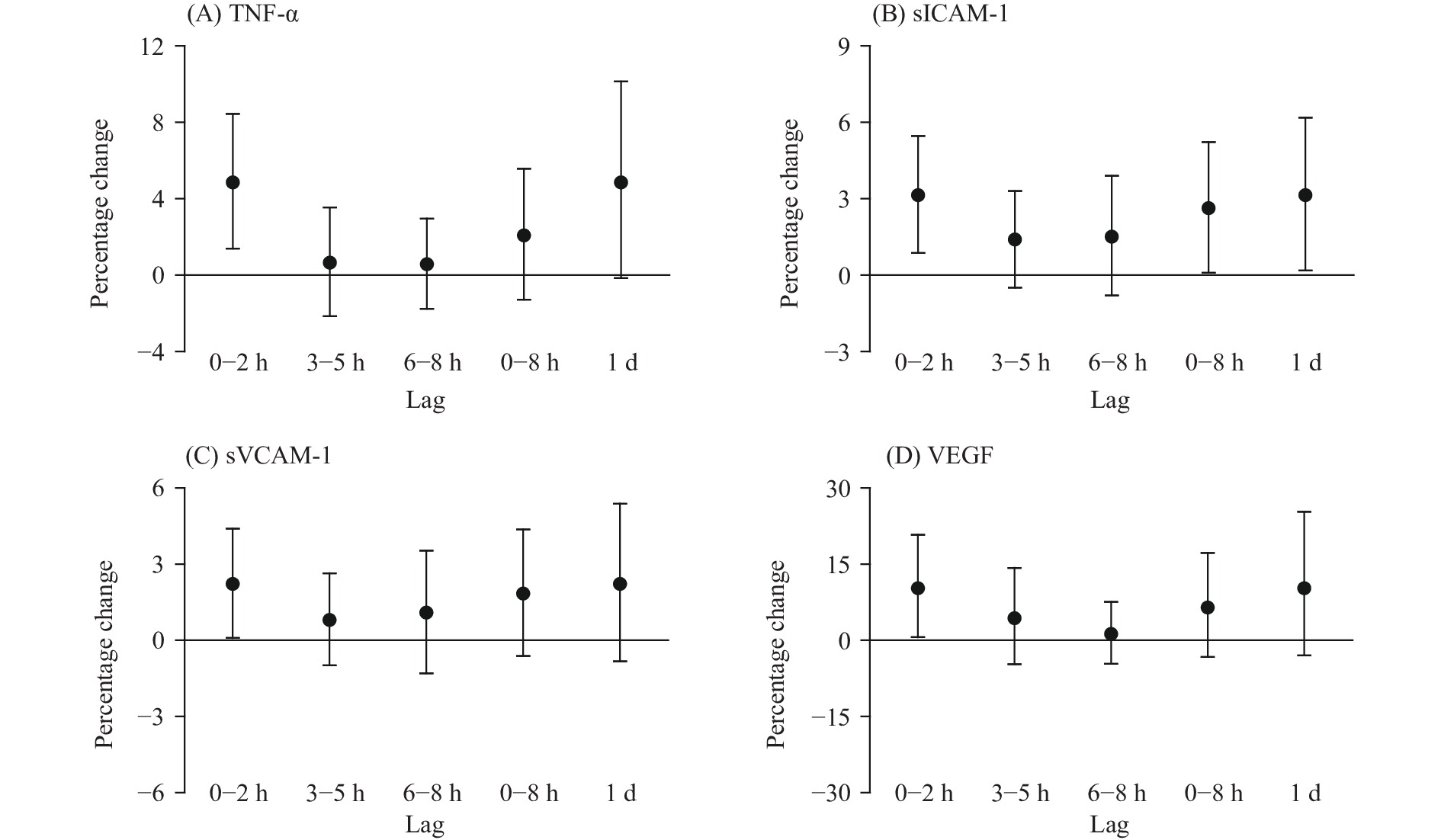 Figure 1.
Figure 1.Percentage changes in TNF-α (A), sICAM-1 (B), sVCAM-1 (C), and VEGF (D) associated with a 10-ppb increase in personal ozone exposure at different lag periods in 40 college students in Shanghai Municipality, China, May–October 2016.
Note: X-axis denotes to the different lag periods; Y-axis denotes the corresponding percentage change in biomarkers.Abbreviations: TNF-α=tumor necrosis factor alpha; sICAM-1=soluble intercellular adhesionmolecule-1; sVCAM-1=soluble vascular cell adhesion molecule-1; VEGF=vascular endothelial growth factor.
HTML
| Citation: |



 Download:
Download: