-
Avian influenza viruses (AIVs) are naturally preserved in waterfowls and sometimes spill over to infect humans and mammalian animals. The H3 subtype is one of the most prevalent influenza virus subtypes in waterfowls. Since 2000, H3 subtype AIVs have been continuously isolated from poultry and wild birds in the mainland of China, which implied a dynamic spread in large-scale geography and multiple species. Combinations of H3 with N1–N8 subtypes were reported, among which H3N2 and H3N8 subtypes predominated. Frequent mutations and reassortments increased the genetic diversity associated with altered virus virulence, transmission, and mammalian adaptation of H3 AIVs, posing a potential threat to animal and human health. This study systematically analyzed the epidemiology, genetic characteristics, and mammalian adaptation-related mutations of H3 subtype AIVs in the mainland of China, facilitating the H3 subtype AIVs research and risk assessment.
Avian influenza virus (AIV) belongs to the Orthomyxoviridae family Influenza virus A genus, packaged with 8 negative-sense and single-strand RNA segments, which encode a total of 14 proteins (1). Based on the antigenic diversity of surface glycoproteins hemagglutinin (HA) and neuraminidase (NA), AIV can be classified into 16 HA subtypes (H1–H16) and 9 NA subtypes (N1–N9) (2-3). Except for the H17N10 and H18N11 subtypes found in bats in recent years, all other subtypes of influenza A virus can be found in wild waterfowls. Therefore, wild waterfowls are known as the natural reservoir and gene pool of influenza A virus, and AIV is considered to be the source of other animal influenza viruses (4). According to the pathogenicity of AIV to chickens, it can be divided into highly pathogenic AIV (HPAIV) and lowly pathogenic AIV (LPAIV) (5). Most avian influenza viruses exhibit low pathogenicity. To date, only partial proportions of H5 and H7 subtype AIVs have developed high pathogenicity, causing high mortality in wild birds, poultry, and even humans.
Among influenza A virus subtypes, the H3 subtype has a wide range of hosts. In addition to circulating in wild birds and poultry, it can infect multiple species of mammals such as humans, pigs, dogs, cats, horses, and even seals (6). The H3 subtype AIV is LPAIV and one of the influenza virus subtypes with the highest isolation rate among ducks (proportion up to 91.76%) (7). Surveillance data showed that the H3 subtype AIV was widely prevalent in domestic ducks in the live poultry markets (LPMs) in China and reassortants were continuously being detected (8-9). Genetically, the HA genes of H3 subtype AIVs could be divided into Eurasian and North American lineages. Both H3 lineages were able to cross the species barrier to infect swine, while Eurasian lineage was identified to infect a wider range of hosts (swine, equine, canine, and human) than North American lineage (swine and seals) (10). In China, only the Eurasian lineage has been detected in domestic poultry (11). The interspecies transmissibility of H3 AIVs historically caused the H3N2 influenza pandemic in 1968 (12), which has posed a threat to human health (13).
Here, we systematically reviewed the current situation of the H3 subtype AIVs in China using surveillance data from 82 published studies and summarized the epidemiological and genetic characteristics and the mammalian adaptation of H3 AIVs in the first 20 years of 21st century in the mainland of China.
-
Since the 1970s, China has reported isolation of AIVs (14). After Hong Kong Special Administrative Region (SAR) reported 18 human infections and 6 deaths caused by HPAIV (H5N1) in 1997 (15), avian influenza surveillance has received global attention for pandemic preparedness. During 2000–2019, AIV surveillance had been conducted in LPMs, poultry farms, and wild birds habitats in the mainland of China. Extensive and continuous active surveillance work included collecting throat and cloacal samples from birds and bird-related environmental specimens such as feces, water, and cage surfaces (11,16-21). Since 2009, surveillance has been intensified in all of 31 provincial-level administrative divisions (PLADs), with a sampling frequency from 2–4 weeks a time to once a year. H3 AIVs were subtyped by RT-PCR or sequencing directly from the samples or AIV isolates.
H3 combinations with multiple NA (N1–N8) subtypes were detected in the mainland of China, dynamically circulating in poultry and wild birds, usually with no apparent illness (22). H3Nx could affect domestic ducks, chickens, geese, pigeons, and quails and mainly circulated in LPMs in China (23-25). From January 2000 to June 2021, 50 studies have reported the detection of H3 subtype AIVs in domestic ducks, including 6 H3 subtype AIVs such as H3N1, H3N2, H3N3, H3N6, H3N7, and H3N8. Overall, 23 articles reported the prevalence of H3 subtype AIVs in domestic chickens. Furthermore, 8 subtypes, H3 combined with N1-N8, have circulated in wild birds, and H3N8 was predominant, with a higher prevalence rate in Anseriformes (20-27). A 4-year surveillance study (2015–2019), including more than 28,000 samples of wild birds from the Qinghai-Tibet plateau, found that H3N8 was the most abundantly detected H3 virus subtype (20).
To summarize the activities of H3 AIVs in the mainland of China, we have counted each H3Nx subtype at the provincial scale each year. For example, if one or more studies reported that H3N2 AIV was found in Heilongjiang Province in 2010, it would be an activity of H3N2 AIV. Since 2000, 214 activities of H3 AIVs have been detected in 26 PLADs in the mainland of China except for the PLADs of Xizang (Tibet), Gansu, Shaanxi, Shanxi, and Hainan (Figure 1A). Surveillance indicated that H3 viruses have become enzootic in domestic ducks in southern China①, where LPMs bring together numerous host species in a high-density setting, creating an ideal environment for viral reassortment and interspecies transmission (23). In eastern China② and northeastern China③, H3 AIVs could affect both domestic poultry and wild birds, except for Fujian, Shandong and Zhejiang provinces where H3 viruses were only detected in domestic poultry (Figure 1A). Previous studies have reported that H3N8 circulated in the Poyang Lake area around Jiangxi Province (18) and the wetlands of Jiangsu Province (28), which are along the wild fowl migratory flyway. In northeastern China, H3 AIVs with multiple NA subtypes (including N2–N8) were detected in wild birds in Heilongjiang Province (Figure 1A), where the complicated bird migration network lies (29). Researchers have continuously detected novel H3N8 subtype AIVs in wild ducks in their habitats such as the Heilongjiang Sanjiang Nature Reserve (30). H3 subtype AIVs were mainly detected in wild birds in northwestern China④, while in southwestern China⑤ they were mainly found in domestic poultry (Figure 1A).
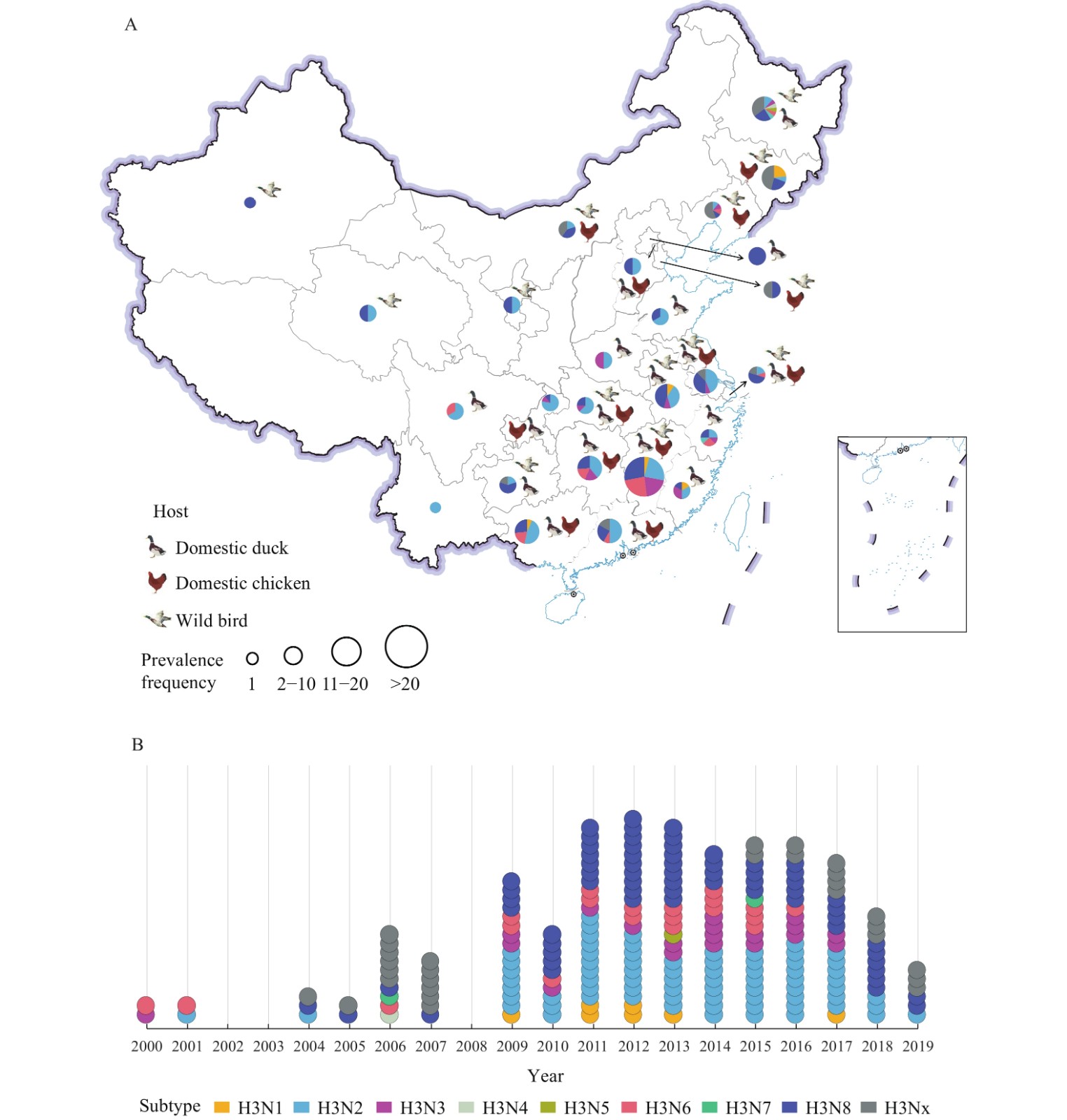 Figure 1.
Figure 1.The spatiotemporal distribution and bird species of H3 subtype avian influenza viruses (AIVs) reported in the mainland of China from 2000 to 2019. (A) The spatial distribution and bird species of H3 subtype AIVs. (B) The temporal distribution of H3 subtype AIVs (according to collection year).
Note: As of June 6, 2021, 214 activities* of H3 subtype AIV have been counted in 26 provincial-level administrative divisions (PLADs) in China since 2000. Each H3Nx subtype is shown as specific color. The diameter of the pie chart (A) represents the number of H3Nx subtype AIV activities. Each circle in (B) represents an activity. Data were accessed from 82 published literatures on PubMed, CNKI, and Wanfang databases, searched up to June 6, 2021.
* Each H3Nx subtype at the provincial scale each year has been counted. For example, if one or more studies reported that H3N2 AIV was found in Heilongjiang Province in 2010, it would be an activity of H3N2 AIV.
In the last decade, increased detection of H3 AIVs was reported throughout the country. Notably, H3N2 and H3N8 have been consistently detected (Figure 1B). An epidemiological survey showed that poultry infection with H3N2 AIV occurred throughout the year in southern China, with higher detection rates in winter and spring and a lower detection rate in summer (31). In the wetlands of northeastern China, virus isolation rate of H3 subtype AIVs varied among seasons, and the highest rate was in autumn, which might due to the migration of wild birds (32).
In summary, H3N2 subtype AIV was the main prevalent subtype in poultry farms and LPMs from China, while H3N8 subtype AIV was widespread among wild birds in this region. Therefore, the following focuses on the H3N2 subtype and H3N8 subtype AIVs.
-
Phylogenetic analysis showed that all of the H3N2 subtype AIVs in China belonged to the Eurasian lineage (11). The HA gene may be derived from viruses circulating in different regions of China and neighbouring countries in East and Southeast Asia (7,17,33), while the NA gene may be derived from other AIVs such as H5 (34-35), H7 (9,36), H9 (23,36), and H11 (34) subtype AIVs in birds in East and Southeast Asia. The prevalence of H3N2 AIVs in birds and frequent reassortments with other AIVs also led to the emergence of various genotypes. Most of the reassortants came from LPMs along the bird migration routes, which suggests that wild bird migration might have a great impact on poultry AIV infection (8,18,23,37). Studies have found that the PA gene of H3N2 subtype AIV of Chinese ducks was highly homologous to the HPAIV H7N9 in Korean wild ducks (33). The H9N2 viruses that were prevalent in poultry in eastern China [Jiangxi (23), Jiangsu (38), Zhejiang (39)] also provided internal genes for H3N2 AIVs. The internal genes of some H3N2 AIVs were closely related to HPAI H5 AIVs, which may suggest reassortment with H5N1, H5N2, H5N3, H5N6, and H5N8 subtype AIVs (8,32,34,40).
-
Molecular epidemiological analysis revealed that the H3N8 subtype AIVs had complex sources. Studies have shown that the HA gene of H3N8 subtype AIV might come from the H3 subtype AIV that is prevalent in poultry in East Asia, Southeast Asia, and Europe (41-42), and the NA gene might come from East Asia, North America, and Europe (43-44). The NA gene of H3N8 subtype AIV could be divided into Eurasian lineage and North American lineage (45). The Eurasian lineage was widely detected in the whole country, while the North American lineage was mainly detected in eastern China (8,11,45), indicating that gene reassortments have occurred between AIVs from the Eurasian lineage and the North American lineage (8,45-47). These events most likely occurred in LPMs and could be transmitted from ducks to chickens (48). The internal genes of H3N8 AIVs were derived from a variety of AIVs from wild birds or domestic ducks (24,49-50), including HPAI H5 viruses (28,43,51-52).
The H3N8 subtype AIVs might be potential gene sources of AIVs causing interspecies transmission. During 2004–2005, researchers detected an H3N8 virus in domestic ducks in Beijing LPMs, of which HA gene was highly similar to the H3N8 virus that caused the 1989–1990 outbreaks in equine populations in northern China (43). In 2018, researchers found that the NA gene of the H3N8 virus isolated from a Guangdong LPM was highly homologous to H10N8 AIV and speculated that the N8 gene originated from the same lineage that caused human infections in 2014 (48).
-
Except for the AIVs of H3N2 and H3N8 subtypes, other AIVs of H3 subtypes were scattered in various regions of China (Figure 1) and had a high degree of genetic diversity and origin. A study indicated that the H5N1 subtype AIV in domestic ducks in Anhui Province and Fujian Province might be involved in the gene reassortment of H3N1 subtype AIV (53). The internal genes of H3N3 might be derived from viruses of waterfowl origin (mainly ducks) in North America (Alaska) and East Asia (Japan and the Republic of Korea) (21-23,39). The HA gene of H3N5 detected in wild birds in Khanka Lake was closely related to poultry-origin AIVs isolated from southern China (21). Studies have shown that H3N6 frequently reassorted with H5N6 subtype AIVs (18,54-55), while PB2 and NP genes were also derived from H9N2 AIV that circulated in the same period (23). Experimental results showed that the novel H3N6 reassortant could effectively replicate in mammalian cells (54). The gene of H3N7 subtype AIV came from a variety of LPAIVs and had greater antigenic differences compared with the H3 subtype AIV strains isolated from previous studies (56).
-
In 1968, a novel H3N2 subtype virus contained genes from human-derived H2N2 influenza virus and avian-derived H3 subtype influenza virus was firstly reported in Hong Kong SAR and caused the pandemic globally (12). It highlighted the pandemic potential of H3 subtype AIVs. Mutations that increased replicative ability and transmissibility of the virus in mammals may facilitate the interspecies transmission (57-59). Previous studies have experimentally demonstrated the relation between some molecular mutations and receptor binding, transmission, replication, pathogenicity, and drug resistance of the H3 subtype AIV in mammals. Here, we have summarized the mammalian adaptation molecular markers in Table 1.
Protein Subtype Mutation Biological effects References PB2 H3N8 K574E Decreased replicative ability in ferrets and transmission ability in mice, decreased RNA polymerase activity in 293T cells (69) T588A Decreased replicative ability in ferrets and transmission ability in mice, decreased RNA polymerase activity in 293T cells (69) E627K,
D701NIncreased replicative ability on mammalian cells and transmission ability among mammals (51) PB1 H3N8 S524G Enhanced RNA polymerase activity in human cells and MDCK cells, increased replicative ability and pathogenicity in mice, enhanced airborne transmissibility between ferrets (45) HA H3N2 G225D Increased HA thermostability and replicative ability on MDCK cells (7) Q226R Increased replicative ability and virulence in ferrets (17) G228S Increased replicative ability and virulence in ferrets (17) H3N8 155T Increased binding capacity of human α2,6-Gal sialic acid receptors (9) NA H3N6 D198N Increased probability of drug resistance, which might reduce the sensitivity of NA inhibitor drugs (such as oseltamivir) (53,70) H3N8 G119P Increased probability of drug resistance, which might reduce the sensitivity of NA inhibitor drugs (such as oseltamivir) (28,70) M1 H3N8 N30D Increased pathogenicity in mice (51) T215A Increased pathogenicity in mice (51) M2 H3N2 V27I Reduce the sensitivity to M2 ion channel blockers (amantadine) (11) S31N Reduce the sensitivity to M2 ion channel blockers (amantadine) (8,11,32,71) NS1 H3N2 D92E Increase probability of viral resistance to interferon, and further experimental verification is needed (67,72) P42S Increased replicative ability and virulence in mice (7) Abbreviation: MDCK=Madin-Darby canine kidney. Table 1. Key amino acid substitutions of H3 subtype avian influenza viruses associated with mammalian adaptation and their biological effects.
Receptor binding is the initial process of virus life circle. AIVs usually show higher binding preference for α2,3-Gal sialic acid receptors (avian-type), and increased binding capacity of α2,6-Gal sialic acid receptors (human-type) indicates increased adaptability in mammals. Studies have shown that H3N2 and H3N8 subtype AIVs could simultaneously bind to both avian-type and human-type receptors (17,22,60). Overall, 2 residues, 226 and 228, of HA of the H3 subtype were well known to be important for the host range restriction (61). Recently, researchers found that residue 155T in the HA protein could enhance the ability of H3N2 subtype AIV to bind to human-type receptors, and the virus could replicate effectively in the lungs and turbinates of mice (9). The G225D mutation of HA could increase the thermal stability of H3N2 AIV, which could probably lead to increased virus replication on Madin-Darby canine kidney (MDCK) cells (7). The H3N2 subtype AIV with Q226R and G228S mutations in HA could enhance the replicative ability and mammalian adaptability in ferrets (17). The Q226L mutation of HA could promote the airborne transmission of the H3N8 virus in ferrets (45).
The RNA polymerase of the influenza virus is composed of three subunits including PB2, PB1, and PA proteins, which are related to the virus’s host-specificity and replication. Dong et al. found that E627K and D701N mutations in the PB2 protein of H3N8 AIV (51) might enhance the adaptability of the virus in mammalian cells and enable AIV to replicate efficiently in mammals (62-65). Another study found that the PB1 S524G mutation of wild bird-origin H3N8 virus could enhance the virulence and fitness for transmission in mammals (45).
Mutations that increase pathogenicity were also found. The N30D and T215A mutations found in the M1 protein could increase the pathogenicity of the H3N8 subtype AIV in mice (51). The residue at 42 of the NS1 protein in the H3N2 subtype AIV changed from P to S, which might increase virus replication and virulence in mice (7).
For drug resistance, the D198N and G119P mutations of NA protein have been identified in the natural isolates of H3N6 and H3N8, respectively. These mutations were likely to cause the H3 subtype AIV with reduced sensitivity of NA inhibitors (28,53). In addition, many studies have found that the V27I and S31N mutations of the matrix protein (M) of H3N2 subtype AIV caused the virus to be resistant to amantadine (8,11,18,32).
-
The H3 subtype AIVs have continuously circulated in wild birds and poultry in the mainland of China. Wild bird migration and live poultry trade play important roles for virus transmission. Frequent gene reassortments were observed between viruses from poultry and wild birds (52,55-66), which increased the genetic diversity of H3 AIVs and contributed genes to other subtype AIVs such as HPAIV H5 (67-68). Despite the relatively improved surveillance studies having been performed in recent years in China, our understanding of the H3 AIVs transmission and evolution is still limited, especially in intercontinental migratory birds. The H3 viruses could acquire mammalian adaptation mutations during replication. Considering the pandemic history and the potential threat to public health, a long-term systematic surveillance of H3 AIV is imperative.
HTML
H3N2 Subtype AIV
H3N8 Subtype AIV
Other H3 Subtypes AIV
MAMMALIAN ADAPTIONS OF H3 SUBTYPE AIV
FootNote
| ① | Including Guangdong Province and Guangxi Zhuang Autonomous Region. |
| ② | Including Anhui Province, Jiangsu Province, Jiangxi Province, Fujian Province, Shandong Province, Zhejiang Province, and Shanghai Municipality. |
| ③ | Including Heilongjiang Province, Jilin Province and Liaoning Province. |
| ④ | Including Qinghai Province, Ningxia Hui Autonomous Region, and Xinjiang Uygur Autonomous Region. |
| ⑤ | Including Sichuan Province and Chongqing Municipality. |
| Citation: |



 Download:
Download:




