-
Influenza is a respiratory illness that infects between 5%–15% of the global population annually during normal seasonal epidemics, and the World Health Organization (WHO) estimates that these infections result in 3–5 million cases of severe illness and about 290,000 to 650,000 respiratory deaths every year (1). Rapid evolution of influenza viruses creates difficulties in recognizing and predicting current and future epidemic threats (2).
Since the March 2020 co-incident with the exponential global spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a dramatic decrease in influenza detection has been observed, despite the testing for influenza continuing at similar levels in many countries (3-4). Influenza is traditionally prevalent in the Southern Hemisphere during April–July 2020, but the positive rate for influenza decreased dramatically from 13.7% in 2017–2019 to 0.06% in 2020 (5). The Northern Hemisphere has also experienced almost no influenza during the typical timing of the 2020–2021 season, compared to around 18%–23% in 2019–2020, the US and EU reporting only 0.15%–0.20% of samples as positive for influenza (6).
Influenza A(H1N1)pdm09, A(H3N2), and influenza B viruses circulated in low numbers and the predominant circulating viruses varied among reporting countries. Since September 2020 influenza activity was mostly reported from countries in the tropics and subtropics and some countries in the temperate zone of the Northern Hemisphere and Southern Hemisphere (7). It is possible that this continuous low-level virus circulation may allow influenza transmission following significant restarting of international travel. Now that we are approaching the influenza season in the mainland of China, it is important that we understand last year’s influenza activity and surveillance results so that we can be better prepared for future influenza outbreaks. Therefore, we analyzed the antigenic and genetic characteristics and antiviral susceptibilities of influenza viruses isolated from the mainland of China in 2020–2021.
-
The Chinese National Influenza Surveillance Network currently includes 410 laboratories and 554 sentinel hospitals. Influenza-like illness (ILI) cases were reported by sentinel hospitals to the Chinese National Influenza Surveillance Information System (CNISIS), and respiratory specimens were collected. Network laboratories determined whether the sample was influenza virus positive using real-time reverse transcription polymerase chain reaction (RT-PCR). Influenza positive specimens were propagated in Madin-Darby canine kidney (MDCK) cells and/or embryonated chicken eggs. The Chinese National Influenza Center (CNIC) received the viruses for further analysis.
Antigenic characterization was based primarily from results of hemagglutination inhibition (HI) assays, and genetic characterization was carried out based on next-generation sequencing. An HI titer less than or equal to 4 times that for the homologous virus was considered to be antigenically similar. Testing of seasonal influenza viruses for resistance to the neuraminidase and endonuclease inhibitors was performed at CNIC using next-generation sequencing analysis, a functional assay, or both. The viruses evaluated were isolated from specimens collected between Week 41 in 2020 (October 5, 2020) and Week 35 in 2021 (September 5, 2021).
-
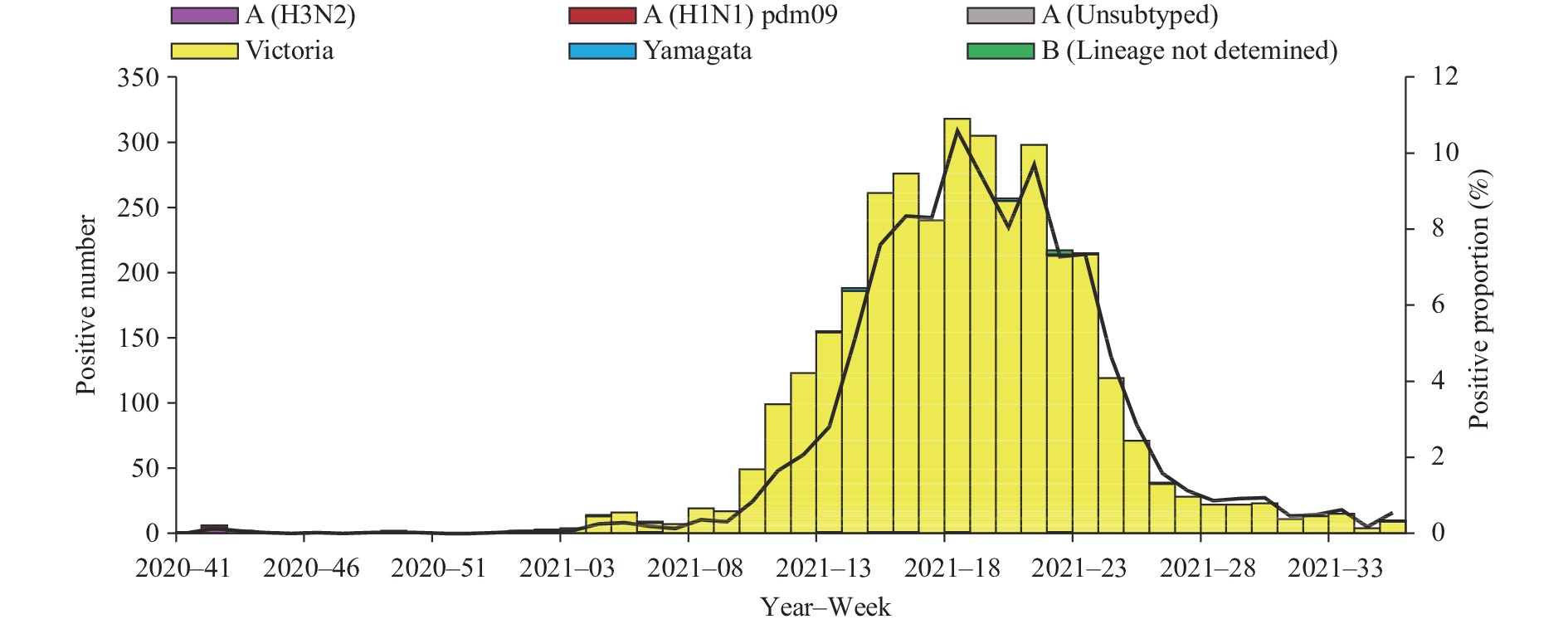
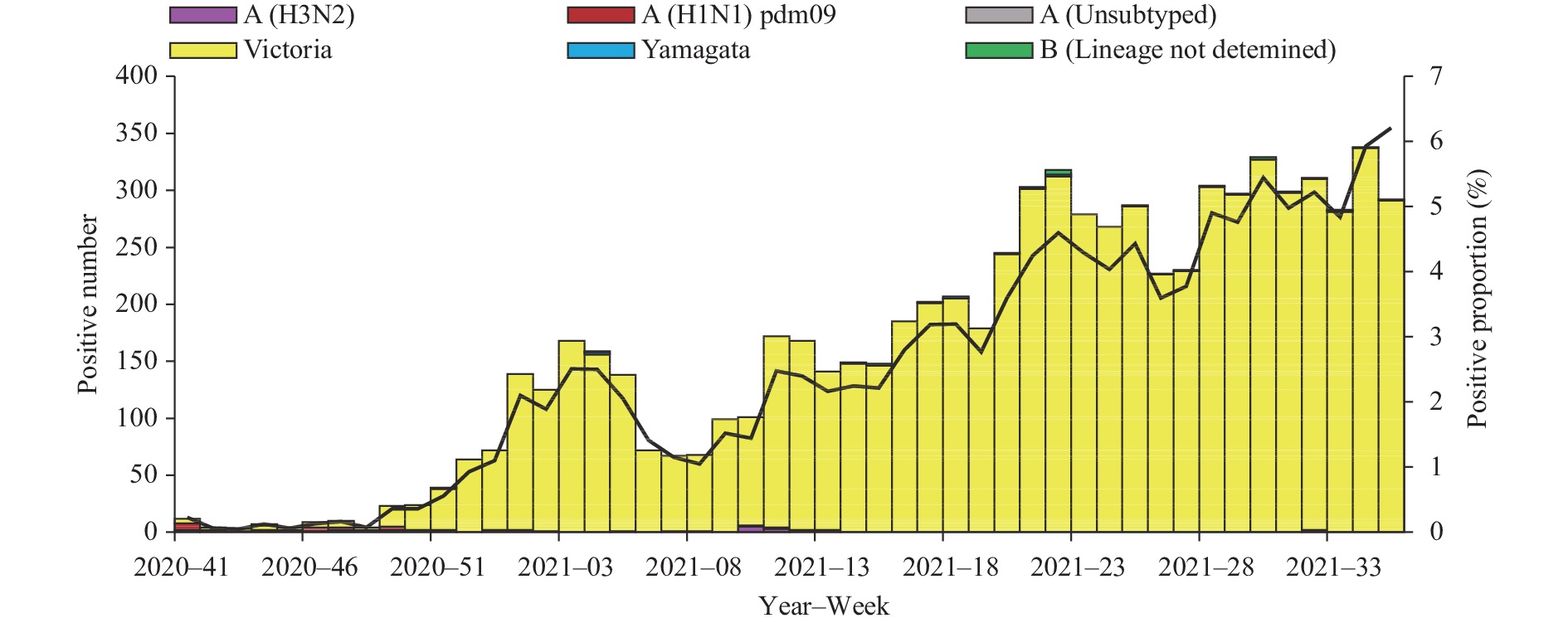
Influenza activity in the mainland of China was below pre-pandemic COVID-19 levels during the period October 5, 2020 to September 5, 2021 (surveillance Week 41 in 2020 to Week 35 in 2021). The percentage of specimens testing positive for influenza each week ranged from 0.1% to 6.2% in southern China and ranged from 0% to 10.6% in northern China. The positivity rate of specimens collected from ILI cases slightly increased starting at Week 49 in 2020 and had been increasing in 2021 in southern China. The positivity rate in northern China began to increase in Week 10 of 2021, peaking at Week 18 of 2021, and declined since then (Figures 1 and 2).
In southern China, the network laboratories tested 308,698 specimens between October 5, 2020 and September 5, 2021; among these specimens, 7,593 (2.5%) tested positive including 79 (1.0%) for influenza A and 7,514 (99.0%) for influenza B. Most of the positive samples (7,299, 96.1%) were collected since 2021. Among the 58 seasonal influenza A positive specimens that were subtyped, 39 (67.2%) were influenza A(H1N1)pdm09 and 19 (32.8%) were influenza A(H3N2). Among the 7,497 influenza B viruses for which lineage was determined, 7,481 (99.8%) belonged to the B/Victoria lineage and 16 (0.2%) belonged to the B/Yamagata lineage (Figure 1).
During the study period, network laboratories in northern China tested 209,219 specimens for influenza; among these, 3,489 (1.7%) were positive for an influenza virus: 19 (0.5%) for influenza A and 3,470 (99.5%) for influenza B. Among the 14 seasonal influenza A viruses subtyped, 7 (50.0%) were influenza A(H1N1)pdm09 and 7 (50.0%) were influenza A(H3N2). Of the 3,465 Influenza B viruses with B lineage results, 3,456 (99.7%) were B/Victoria and 9 (0.3%) were B/Yamagata (Figure 2).
Two A(H1N1)pdm09 viruses were antigenically characterized by HI tests, and one virus was well inhibited by ferret antisera raised against A/Guangdong-Maonan/SWL1536/2019, the egg-propagated reference virus representing the A(H1N1)pdm09 component of the 2020–2021 Northern Hemisphere influenza vaccines (7). One of the viruses was well inhibited by ferret antisera raised against A/Victoria/2570/2019, the egg-propagated reference virus representing the A(H1N1)pdm09 component for the upcoming 2021–2022 Northern Hemisphere influenza vaccines (8). Phylogenetic analysis of hemagglutinin (HA) gene segments determined that 1 belonged to genetic subclade 6B.1A5A with HA amino acid substitutions N129D, T185I, and N260D; the other one belonged to group 6B.1A5A2 with HA amino acid substitutions K130N, N156K, L161I, V250A, and E506D, which evolved from subclade 6B.1A5A.
Antigenic characterization of the 1,819 B/Victoria lineage viruses were conducted using HI tests. A total of 1,141 (62.7%) B/Victoria lineage viruses were not recognized well by the ferret antisera raised against B/Washington/02/2019, the egg-propagated reference virus representing the B/Victoria lineage component of the 2020–2021 and 2021–2022 Northern Hemisphere influenza vaccines (7-8). Among the 797 viruses HA gene segments sequenced and phylogenetically analyzed, all belonged to genetic clade 1A, nearly all belonged to subclade 1A.3, which shared a three amino acid deletion in HA (positions 162–164) and the substitution K136E, and in which, 96.7% belong to group 1A.3a characterized by HA amino acid substitutions N150K, G184E, N197D, and R279K, and was further divided into subgroup 1A.3a1 (520, 65.2%), which had additional HA amino acid substitutions V220M and P241Q, and 1A.3a2 (251, 31.5%), which had HA amino acid substitutions A127T, P144L, and K203R.
No A(H3N2) and B/Yamagata lineage viruses were isolated and available for characterization during the study period.
Of the 2 A(H1N1)pdm09 viruses collected in the mainland of China, neither showed evidence of reduced inhibition by oseltamivir and zanamivir, nor to baloxavir. Of the 1,258 influenza B/Victoria lineage viruses screened for neuraminidase inhibitors susceptibility, all showed normal inhibition by oseltamivir and zanamivir. A total of 797 B/Victoria lineage viruses were screened for susceptibility to the endonuclease inhibitor, baloxavir, by genetic analysis, and none showed evidence of reduced susceptibility.
-
In the mainland of China, circulation of influenza virus was disrupted during the COVID-19 pandemic. During 2020, influenza virus circulation showed historically low levels (9). In 2021, influenza continues to circulate at low levels but higher than the equivalent period since March 2020 (9).
The WHO meets twice a year to recommend influenza vaccines. In late February 2021, the WHO issued its recommendations for the 2021–2022 Northern Hemisphere influenza vaccines. Compared to the 2020–2021 Northern Hemisphere influenza vaccine components, the A(H1N1)pdm09 and A(H3N2) vaccine components were updated, except for the B/Yamagata and B/Victoria lineage (7-8).
Influenza B/Victoria lineage viruses were predominant nationwide during the study period. Significant genetic diversity was seen in circulating viruses of the B/Victoria lineage. The viruses in the 1A.3a group were the most prevalent in the mainland of China. This 1A.3a HA group further diversified into 2 subgroups that 1A.3a1 had substitutions of either V220M and P241Q, which were predominant and seen almost exclusively in China, or 1A.3a2 with additional amino acid substitutions A127T, P144L, and K203R, for which the proportion has been increasing in recent months. Subgroup 1A.3a2 shows further genetic differentiation, with additional HA amino acid substitutions found in viruses from other global regions (10). The majority of group 1A.3a viruses showed reductions in inhibition by post-infection ferret antisera raised against egg-propagated 1A.3 viruses, such as B/Washington/02/2019 (2020–2021 and 2021–2022 Northern Hemisphere influenza vaccine components).
Only 2 A(H1N1)pdm09 viruses were available for antigenic analysis during this period. Ferret antisera raised against egg-propagated A/Victoria/2570/2019 (2021–2022 Northern Hemisphere influenza vaccine components), which belonged to phylogenetic group 6B.1A5A2, recognized the virus within 6B.1A5A2 virus well but showed poor recognition of 6B.1A5A virus. The global 6B.1A5A2 viruses increased rapidly in the early 2020s until it reached similar proportions to 6B.1A5A1, but 6B.1A5A1 viruses dominant from September 2020 and 6B.1A5A2 viruses were recently detected (10).
No A(H3N2) and B/Yamagata lineage viruses were available for characterization during the study period. Influenza A(H3N2) viruses were reported in most regions, including Africa, Asia, North America, Oceania and Europe, but the clades of A(H3N2) viruses circulating varied among reporting countries. The largest proportion of 3C.2a1b2a2 viruses with HA amino acid substitutions K83E, Y94N, T131K, Y159N, T160I, L164Q, G186D, D190N, F193S, and Y195F in this period were found in South Asia, Southeast Asia, the Middle East, East Africa, Oceania, North America, and Europe (10). There are no B/Yamagata lineage viruses isolated globally with dates after March 2020 (10).
Based on our analysis, all tested viruses for susceptibility to therapeutics were susceptible to neuraminidase and endonuclease inhibitors. Anti-influenza drugs were used as an effective means of post-exposure prophylaxis and treatment of influenza virus infection. Continuous monitoring of circulating influenza viruses for antiviral resistance is still important.
Because of the common transmission route between SARS-CoV-2 and influenza, the same protective behaviors can greatly limit the spread of influenza (11-13). In the mainland of China, B/Victoria lineage viruses continued to be identified, but the diversity of types/subtypes co-circulating has decreased compared to recent seasons. Globally, few B/Yamagata lineage viruses have been found during the COVID-19 pandemic. The surveillance data presented here are a reminder that the low level of influenza circulation since early 2020 may not necessarily persist into the upcoming influenza season space. Given the long absence of sustained natural exposure to influenza viruses, the reduction in influenza virus transmission over the past year may affect the severity of the upcoming influenza season. Lower levels of population immunity, especially in young children and the elderly, may portend more widespread illness and potentially more severe epidemics when the influenza viruses re-emerge (14-15). The lack of influenza circulation in the previous season will reduce our confidence in the strains that may circulate in the next season and may result in mismatch between the vaccine and circulating strains. Maintaining surveillance and outbreak response is essential to track geographic spread of the virus and its variation characteristics so that the vaccine remains optimized for circulating influenza viruses, and influenza surveillance should continue to be strengthened.
HTML
| Citation: |



 Download:
Download: