-
In December 2019, after the coronavirus disease 2019 (COVID-19) outbreak in China, the Chinese government implemented a series of strong measures including the introduction of COVID-19 into the mandatory reporting of infectious diseases, the lockdown of Wuhan, and the quarantine of all inbound passengers followed by isolation at designated sites for 14 days; under no circumstances were any foreign crewmembers of cargo ships allowed ashore. By the middle of April, China had successfully interrupted the virus’ transmission in the community.
COVID-19 has since been largely controlled in China. However, several outbreaks or clusters of COVID-19 cases have occurred and have been linked to importation of cases or from suspected virus-contaminated imported products. Several outbreaks were suspected to have occurred due to the handling of cold chain seafood by the first infected case, especially with outbreaks in Beijing’s Xinfadi and Dalian City (1-3). After epidemiological investigation, the full-length viral genome in those outbreaks could be traced back to SARS-CoV-2 contamination of the outer packaging of cold chain products. However, the most important evidence, the isolation of the live virus, was not obtained.
Presently, there is still no consensus on whether the outer packaging of the cold chain products can carry live viruses and cause virus transmission. The World Health Organization (WHO) and Food and Agriculture Organization (FAO) of the United Nations believed that the outer packaging of cold chain products could not carry live viruses and cause virus transmission (4-5). However, China’s key ports of entry by air, land, and sea have faced outbreaks and challenges related to the COVID-19 epidemic. Qingdao, a major port in eastern Shandong Province and China’s second largest foreign trade port, started active monitoring using nucleic acid testing on 14-day intervals in August 2020 for the populations at high risk and those that process, sell, and handle frozen aquatic products. On September 22 and 24, 2 cold chain dock workers from the same company screened positive for SARS-CoV-2; these cases became the first reported in 151 consecutive days in Qingdao City. To identify the source of infection and evaluate the risk of virus transmission from the contaminated outer packaging of cold chain products, an in-depth field epidemiological investigation was conducted. Full-length viral genome sequencing, real-time reverse transcription polymerase-chain reaction (rtRT-PCR), and virus isolation were carried out.
HTML
-
Since August 11, 2020, high-risk individuals in Qingdao city, including people working in frozen seafood processing, sales, and shipment chains, were routinely tested every 14 days for SARS-CoV-2 nucleic acids. By September 8, the 573,107 residents had tested negative 3 times for SARS-CoV-2 nucleic acid by rtRT-PCR. Throat swab samples that were collected from the identified 2 dock workers between September 22 and 24, 2020 tested positive with cycle threshold (Ct) values targeting the ORF1ab gene of 37 and 23 and the N-gene of 37 and 25. IgM/IgG OD value of Dock Worker 1 were <1/<1, 2.17/<1, and 8.26/3.54, respectively; IgM/IgG OD value of Dock Worker 2 were <1/<1, 2.55/<1, and 3.22/6.77 on September 25, October 2, and October 13, respectively. This revealed that the suspected exposure period was between September 8 and 23, 2020. Throat swabs from 232 close contacts of the two dock workers, including 69 dock workers who participated in the unloading of Transport Ship K (Ship K), as well as 2,019 residents who were placed under home quarantine tested negative for SARS-CoV-2 nucleic acid. The results of the serum antibody tests were all negative.ck workers who participated in the unloading of Transport Ship K (Ship K), as well as 2,019 residents who were placed under home quarantine tested negative for SARS-CoV-2 nucleic acid. The results of the serum antibody tests were all negative.
Between September 8 to 23, 2020, the 2 asymptomatic infected dock workers had no history of living in high-risk areas in China and had no contact with patients from high-risk areas or patients with unexplained fever. There were no imported cases in their communities for 55 days, and neither dock worker had contact history with people returning from overseas. During the period of likely exposure, the two infected dock workers only unloaded frozen cod from abroad on Ship K.
After Ship K docked arrived at the port in Qingdao City, 71 dock workers unloaded 2,100 tons of frozen cod from September 18 to September 20, 2020. The 71 dock workers were divided into 3 working groups, and each group worked for 10 hours. The 2 infected dock workers handled frozen cod from 2 different storage cabins from 20∶00 on September 19 to 06∶00 on September 20, 2020.
To identify the risk factors in handling frozen cod on the Ship K, a retrospective cohort study was conducted using a structured questionnaire. All 71 dock workers who carried the cold chain products of Ship K were interviewed by telephone survey to investigate the crewmembers’ contact history and behavioral factors during loading of the frozen cod on Ship K. The risk behaviors of the two infected dock workers included smoking, wearing masks with their noses exposed, and improper washing of hands during the boarding of the ship and loading and unloading of frozen cod on Ship K. Although all 71 dock workers had habits of smoking, only 3 dock workers smoked during the loading and unloading of the frozen cod. The risk of infection among the smoking dock workers while handling frozen cod was 67% (2 of 3), and none of the 68 dock workers who did not smoke during loading and unloading were infected. The difference between the two groups was statistically significant (P=0.001).
-
About 90 crewmembers boarded Fishing Trawler V (Trawler V) from July 2020. We reviewed the SARS-CoV-2 nucleic acid test results of the crewmembers of Trawler V. The 90 crewmembers were not tested for SAS-CoV-2 nucleic acids. The packaged frozen cod were transferred from Trawler V to Ship K on September 3, 2020. The crewmembers of Trawler V fetched the cod from their cold storage, transferred the cod to Ship K via cranes, and then froze (−18 °C) the cod on Ship K.
The 23 crewmembers of Ship K repeatedly tested negative for SARS-CoV-2 nucleic acids including before arrival in Qingdao (tested on August 18 and September 9, 2020), upon arrival in Qingdao (tested on September 16, 2020), and upon arrival in Busan, the Republic of Korea (tested on September 21, 2020). Furthermore, there was no contact history between the crew of Ship K and the frozen cod (Figure 1).
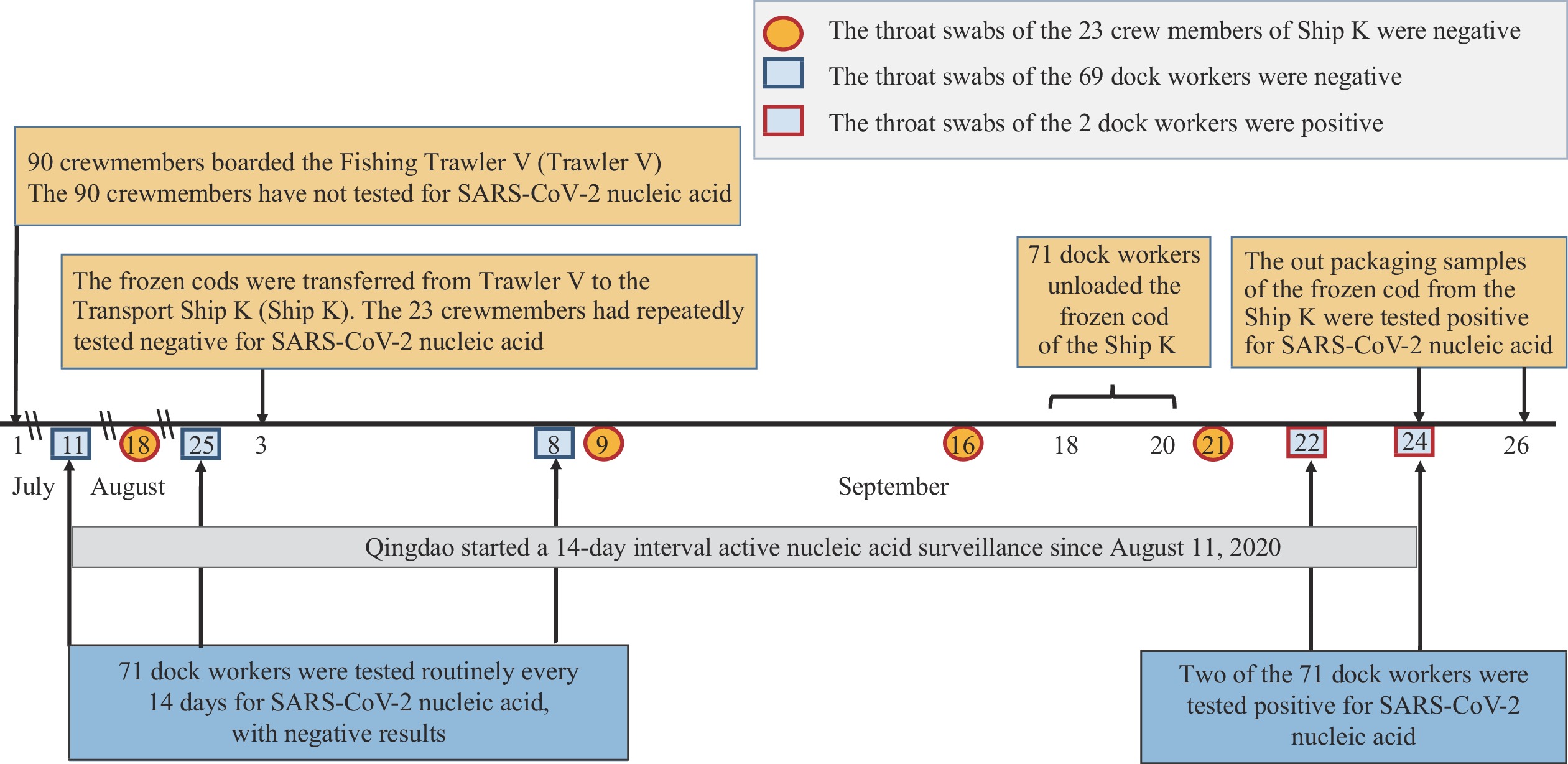 Figure 1.
Figure 1.Timeline of SARS-CoV-2 transmission from contaminated cold chain products to humans — Qingdao City, Shandong Province, China, September 2020.
Note: A timeline of Fishing Trawler V being at sea, Transport Ship K being at sea, the transfer of cod from Trawler V to Ship K, routine SARS-CoV-2 screening of transport Ship K’s crew, unloading at Qingdao port, positive tests being found among 2 dock workers, and testing of the cod packages.The 71 dock workers were interviewed individually. It was determined that the distance between the dock workers and the crew of Ship K was more than 10 meters. The dock workers did not have direct contact with the crew of Ship K during the cod handling process, and they all wore protective clothing, medical masks, and disposable gloves. Since the crews of Ship K did not have any evidence of SARS-CoV-2 infection, the origin of SARS-CoV-2 contamination of the packaging of frozen foods was highly likely from the crewmembers of Trawler V.
-
To assess contamination and increased viral load in the environment, a two-stage sampling method was used to sample the outer packaging of the cod from Ship K. All nucleic acid tests were completed within 24 hours after environmental sample collection. The rtRT-PCR Ct value of ORF1ab and N gene target ≤40 was positive.
First-stage sampling was conducted on September 24, 2020 to determine which outer packaging was positive. In the cold storage of the frozen cod, 70 pallets (about 63 cartons of cod per pallet) were selected for sampling. The four surfaces of each pallet were sampled separately. Each smear sample that was pre-wetted with the viral transport medium (VTM) was smeared at five positions on each surface in one direction. A total of 420 smear samples were collected, and each swab was stored separately in a virus sampling tube containing 3 mL of VTM; the tail of the swab was then discarded, and the tube cap was screwed tightly for transportation in an ice box. Among the 420 smear samples, 51 were positive for SARS-CoV-2 nucleic acids.
To collect samples with high viral loads for virus gene sequencing and virus isolation, second-stage sampling was carried out on September 26, 2020 on the first SARS-CoV-2 nucleic acid positive 8 pallets. The 8 pallets were placed on the platform outside the cold storage by a forklift truck. Smear samples from all five outer surfaces of each carton of cod (for each of the five pre-wetted swabs) were placed in a virus sampling tube containing 3 mL of VTM (without virus inactivation), and the tube caps were screwed tightly for transportation in an ice box. A total of 499 smear samples were collected, and 51 were positive for SARS-CoV-2 nucleic acids.
-
Overall, 2 case samples and 11 frozen cod samples with positive nucleic acid results (Ct value <32) were studied with whole genome sequencing. The viral RNAs were extracted directly from the swab samples with the QIAamp Viral RNA Mini Kit (QIAGEN, Germany), and rtRT-PCR assay was used to detect SARS-CoV-2 nucleic acid. For genome sequencing, the libraries were prepared using the Nextera XT Library Prep Kit (Illumina, San Diego, CA, USA), and the resulting DNA libraries were sequenced on MiSeq platforms (Illumina, San Diego, CA, USA) using a 300-cycle reagent kit. Mapped assemblies were done using the SARS-CoV-2 genome (accession number NC_045512) as a reference. Variant calling, genome alignments, and sequence illustrations were generated with CLCBio software (QIAGEN, Germany).
A total of 2 throat swabs samples from the 2 dock workers and 11 environmental samples tested positive for SARS-CoV-2 nucleic acids with Ct values <35. After high-throughput sequencing, a total of 9 SARS-CoV-2 genomes were obtained through high-throughput sequencing from the 2 infected dock workers, 7 outer packaging samples, 1 secondary sample taken from the outer packaging, and the isolated virus strains (the other 10 genomes all came from uncultured samples). Compared with the Wuhan reference strain (NC_045512) sequence, these 11 whole genomes had 12–14 nucleotide mutation sites and shared 12 nucleotide mutation sites. Among them, 7 shared mutation sites (C241T, C3037T, C14408T, A23403G, and GGG28881-28883AAC) were consistent with the characteristics of lineage B.1.1 (Figure 2) (6–7). The other 5 shared mutation sites (C3743T, C3773T, C5170T, A23299G, and G23755T) that were unique to SARS-CoV-2 were identified in Qingdao (Figure 3).
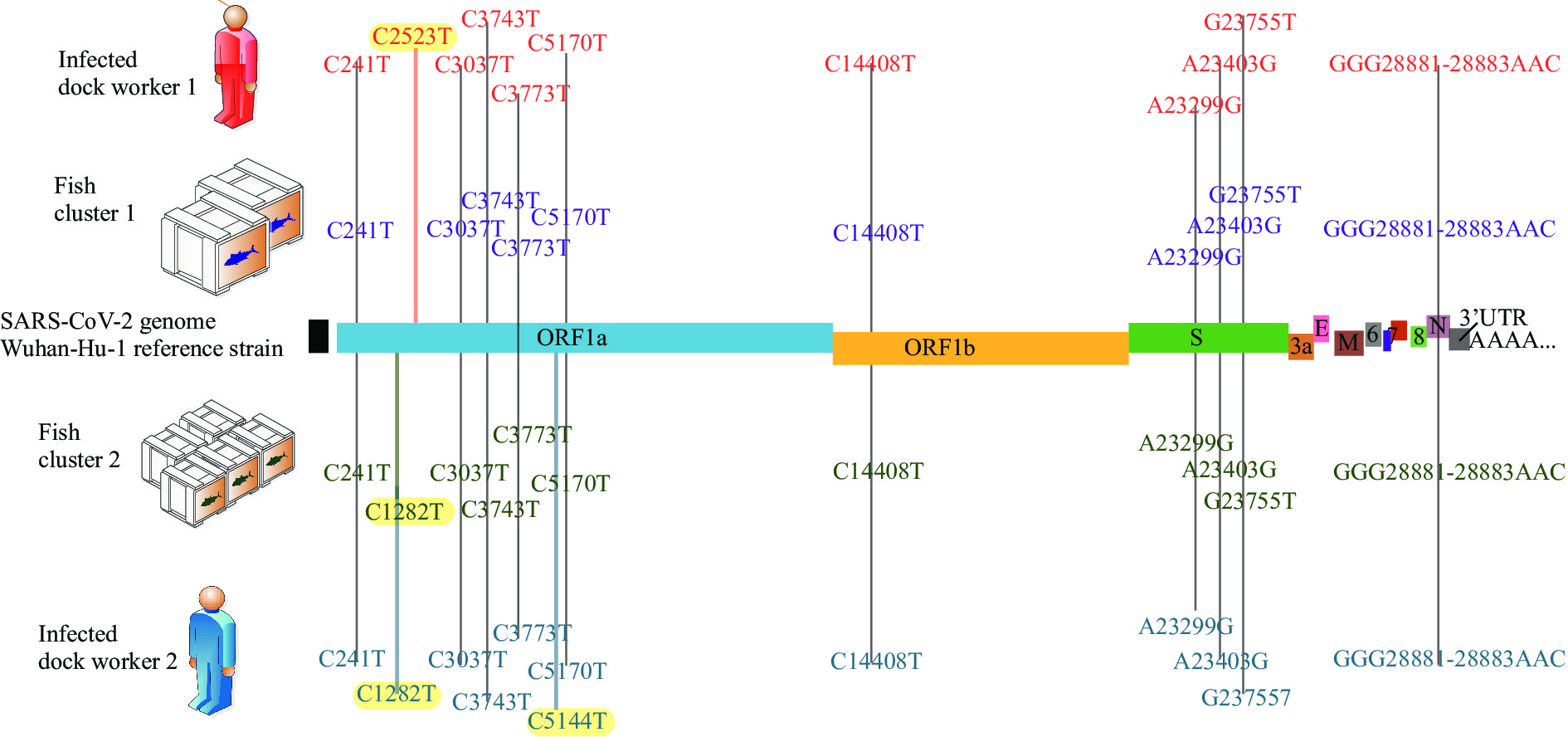 Figure 2.
Figure 2.A multiple genome alignment of the SARS-CoV-2 genome sequences obtained from the two dock workers and from the environmental swab samples with reference strain (Strain Wuhan-Hu-1).
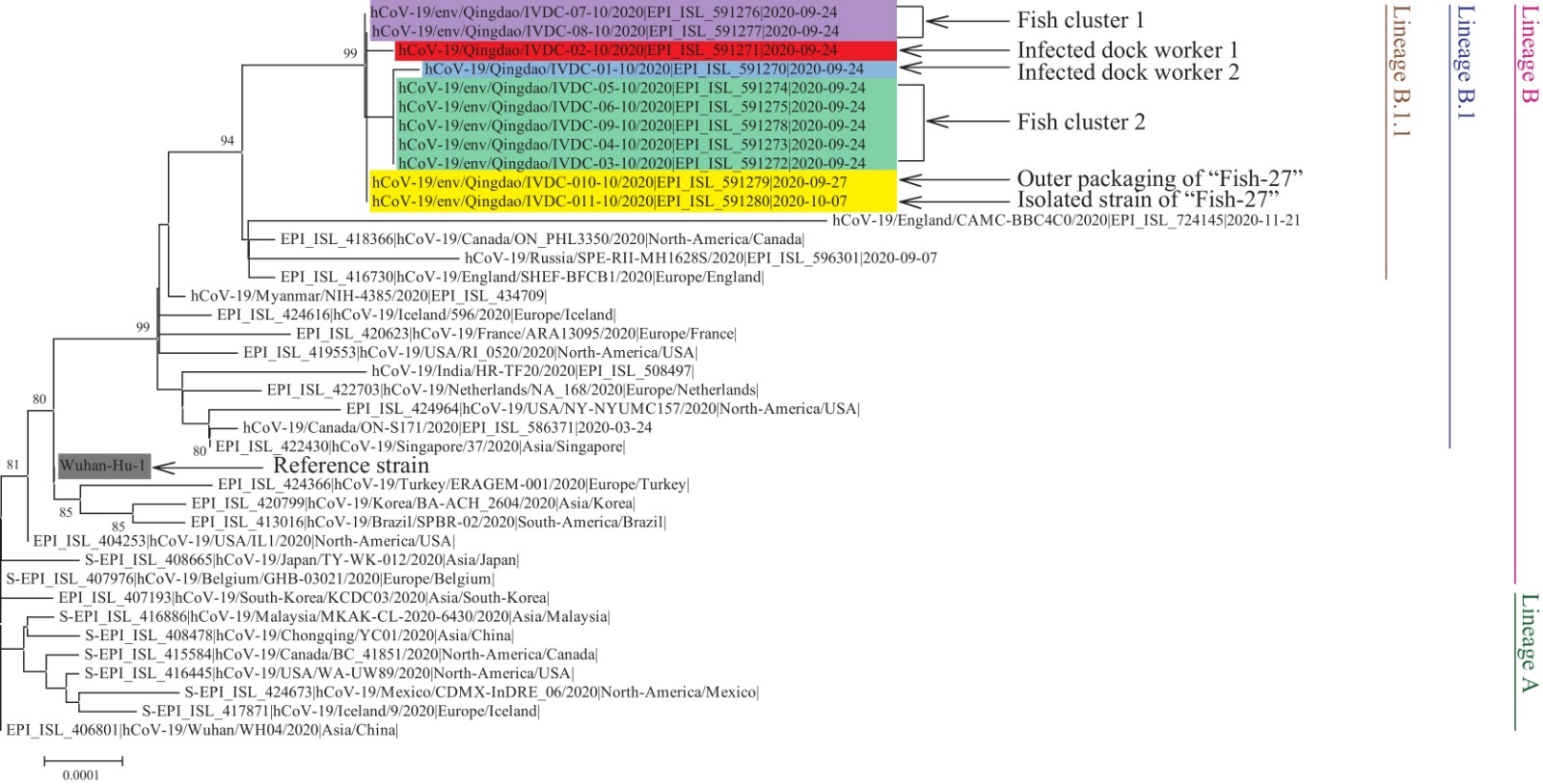 Figure 3.
Figure 3.Phylogenetic analysis of SARS-CoV-2 based on full-length genome sequences.
Note: The SARS-CoV-2 strain that caused the Qingdao outbreak belonged to lineage B.1.1. The sequences derived from the “Fish 27”, Fish clusters 1 and 2, and infected dock workers 1 and 2 were highlighted in shades of yellow, purple, green, red, and blue, respectively. The lineages of SARS-CoV-2 were marked and colored on the right.In addition to the 12 shared nucleotide mutation sites, the strain from dock worker 1 had 1 nucleotide mutation (C2523T) and that from dock worker 2 had 2 additional nucleotide substitutions (C1282T and C5144T); 5 outer packaging-related strains shared 1 unique nucleotide mutation (C1282T) with the strain from dock worker 2, and no additional nucleotide substitutions were found in the remaining 2 outer packaging strains (Figure 4). Therefore the viral genome sequence of dock worker 1 shared 12 mutations and added one specific mutation (C2523T) compared to the 2 viral genome sequences from Fish cluster 1, while the viral genome sequence of dock worker 2 shared 13 mutations and added one specific mutation (C5144T) compared to the 5 viral genome sequences from Fish cluster 2. It was preliminarily determined that the 2 dock workers were infected with SARS-CoV-2 by loading and unloading the frozen cod with contaminated outer packaging.
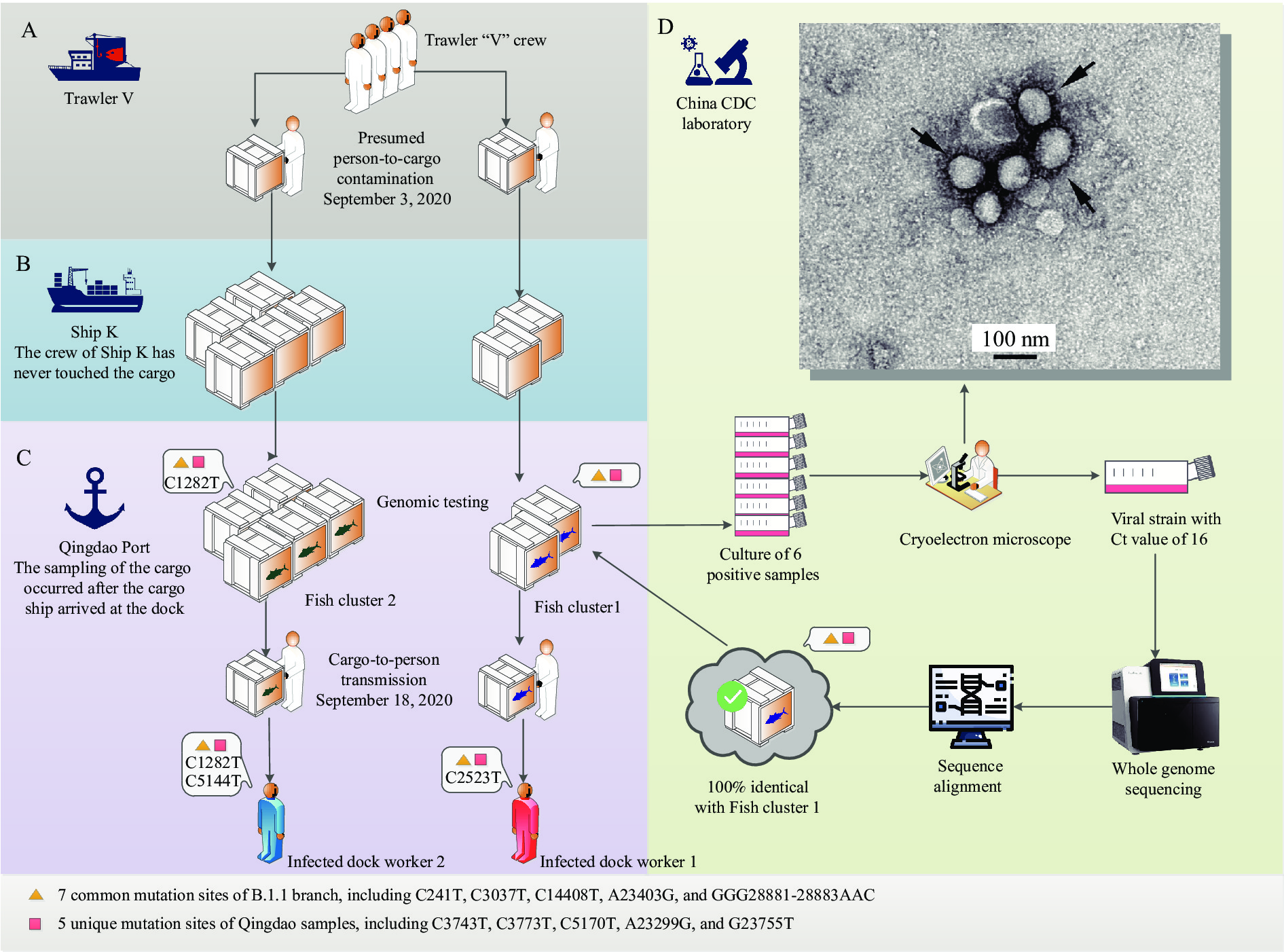 Figure 4.
Figure 4.Schematic diagram of two transmission chains of SARS-CoV-2 from frozen product packaging to humans.
(A) Person-to-cargo transmission of SARS-CoV-2 in Trawler V where the crew of Trawler V loaded the combined cargo. (B) Long distance transportation of cold chain seafood package contaminated with SARS-CoV-2 where the crew of Ship K had no contact with the cargo. (C) Cargo-to-person transmission of SARS-CoV-2 with new substitutions in Qingdao Port where the dock workers unloaded the combined cargo and then the 2 dock workers tested positive; the fish cargo was tested after it was unloaded. (D) Viral isolation and electron microscopic examination. Live virus was successfully isolated, full-length genome sequence of the live virus was 100% identical to the sequence from the swab smearing samples of cold-chain seafood packaging (Fish Cluster 1). The upper right picture shows the electron microscope image of the live virus. The orange triangle indicates 7 common mutation sites of lineage B.1.1 of SARS-CoV-2, including C241T, C3037T, C14408T, A23403G, and GGG28881-28883AAC. The red square indicates five unique mutation sites of SARS-CoV-2 identified in Qingdao, including C3743T, C3773T, C5170T, A23299G, and G23755T. -
During the second-stage sampling, 6 samples with high viral loads (Ct values 25, 28, 30, 31, 31, and 32) collected from Fish Cluster Pallet 1 were obtained for further viral isolation. The cell tube inoculated with specimen “Fish-27” with a Ct value of 25, showed obvious cytopathic effects after the second passage (Figure 4). Typical coronavirus-like particles were observed under cryoelectron microscope, which indicated live, infectious SARS-CoV-2 was isolated from specimen “Fish-27”. The sequencing results showed that the viral genomes from the specimen “Fish-27” and the corresponding strain shared 100% nucleic acid identity, and it showed that the viral sequence only had 12 nucleotide mutation sites, not including C1282T, C5144T, or C2523T. The sequencing match also suggested that the positive virus culture was not a result of contamination.
-
The 11 full-length genomic sequences detected in this study have been deposited to GISAID database under accession numbers EPI ISL 591270 to EPI ISL 591280.
-
This study confirmed that imported cold chain products were contaminated with relatively high loads of SARS-CoV-2 through the excrement of Fishing Trawler workers infected with the SARS-CoV-2. After the contaminated cold chain products transported from Fishing Trawler to the transport ship, the SARS-CoV-2 can spread across countries and regions over long distances through the international marine fishery trade. This was similar to close contact transmission in families and medical institutions following SARS-CoV-2 contamination of external environments. This has brought about serious public health challenges to countries and regions that have successfully interrupted local SARS-CoV-2 transmission.
First, when contaminated packages have high viral load, the virus can survive for weeks to months (July to September being the longest recorded) at low temperatures, and the virus may still infect employees after being in cold storage for months or years, resulting in silent transmission of a “relatively old virus”. Second, it is strongly suggested that the management of the cold chain process should be strengthened globally to prevent SARS-CoV-2 contamination during fish and meat production. Regular screening for SARS-CoV-2 nucleic acids in high-risk population is necessary to identify infected persons at an early stage. This will prevent both cold chain contamination by SARS-CoV-2 and potential silent transmission. Third, this study suggested that SARS-CoV-2 can survive in humid, low temperatures and high-salt environments, which is an important scientific issue. Thus, it is necessary to conduct further research on the influence of environmental factors, such as pH, salinity, temperature, and humidity, on the survival or transmission of the SARS-CoV-2. However, it is important to note that while further study and surveillance is possible, maintaining these efforts may be difficult given the infrequency of these outbreaks relative to the massive amount of trade in Chinese ports.
Epidemiologic and Laboratory Information from the Dock Workers
Epidemiologic and Laboratory Results for Transport Ship K
Environmental Investigation and Sampling
SARS-CoV-2 Detection and Genome Sequencing
Isolation of SARS-CoV-2
Data Availability
Public Health Response
-
Since the outbreak of COVID-19, SARS-CoV-2 nucleic acids have been detected in the outer packaging of several cold-chain seafood products imported from abroad (8). It has long been suspected that SARS-CoV-2 contamination of the outer packaging of cold-chain seafood products could lead to infection among employees handling the outer packaging. This may be a hidden route of transmission due to the long-distance transportation of the cold-chain seafood products.
SARS-CoV-2 was found to be more stable on plastics and stainless steel than on copper and cardboard materials, and live virus was detected within 72 hours after application to these surfaces (9). Therefore, frequent exposure to contaminated surfaces in public places is a potential route of SARS-CoV-2 spread. Swabs were collected from the outer package of cold chain products, but the swabs collected from the seafood itself were negative, indicating that the products were likely not infected with SARS-CoV-2.
Furthermore, after the infected dock workers had been sent to Qingdao Chest Hospital for further investigation and treatment, an outbreak alarm was triggered on October 11, 2020 for an additional 12 cases, and initial epidemiological evidence suggested in a previous report that all 12 cases were linked to Qingdao Chest Hospital (10). They did not have contact with other people who were later found to be infected, but their chest computed tomography (CT) scans were done in the same CT suite that was visited by a hospital patient and a nursing assistant who later tested positive. After investigating the 12 cases and their close contacts, the 2 dock workers and their close contacts were not found to have any temporal and spatial links outside Qingdao Chest Hospital. The overseas frozen cod that caused the infection of two dock workers had also been sealed in the customs warehouse without entering the country. This evidence suggested the transmission in a ward of Qingdao Chest Hospital, forming a cluster epidemic. Fortunately, due to the timely discovery of the cases, this incident did not result in large-scale community transmission.
It should be emphasized that, so far, no cases of consumer infection due to exposure to contaminated cold chain seafood have been found, and the risk of consumer infection is, therefore, extremely low. In addition, the contamination of cold chain products by SARS-CoV-2 is an accidental event, and it is difficult to identify contaminated products by screening. Therefore, it is suggested that regular screening and detection should be strengthened (and testing should occur every 14 days). Monitoring of SARS-CoV-2 nuclear acid levels in employees of imported seafood cold chain products (especially workers engaged in port operations and seafood processing enterprises), should also be implemented for early detection of infected persons and early interruption of transmission.
We also noticed that the two dock workers infected with SARS-CoV-2 at the wharf took off their masks and smoked, while none of the other dock workers who had been wearing their masks were infected with SARS-CoV-2. This shows that SARS-CoV-2 can be transmitted from the outer packaging of the cold chain to the dock workers; however, infection can likely be avoided by using proper protection.
The major limitation of this study was that the symptoms and SARS-CoV-2 infection status of the crewmembers of Trawler V were unknown. If these crewmembers aboard Trawler V were indeed infected, then the transmission chain could be readily established.
It is particularly important that the international community should pay close attention to SARS-CoV-2 transmission by cold chain, build international cooperative efforts in response, share relevant data, and call on all countries to take effective prevention and control measures to prevent SARS-CoV-2 contamination in cold chain food production, marine fishing and processing, transportation, and other operations. At the same time, it is necessary to strengthen the health and epidemic prevention knowledge of employees and those in related occupations who are engaged in handling the outer packaging of imported seafood cold chain products. Formulation of standards for equipment and facilities related to the working environment of imported cold chain food and personal protection standards should be improved. It is also suggested that staff at the port should be included as a high-risk population for emergency vaccination.
| Citation: |




 Download:
Download:




