2022 Vol. 4, No. 49
Antibiotic resistance (AR) is a serious public health threat worldwide. However, the AR and antibiotic resistance genes (ARGs) data from West Africa, especially from Sierra Leone, are limited.
The study revealed ARGs’ common dissemination, and multiplex antibiotic resistance genes in one sample. Genes blaNDM and blaOXA-48-like were first discovered in Sierra Leone.
Basic information is provided for AR research and surveillance and highlights that effective AR surveillance among diarrhea patients is necessary for Sierra Leone and West Africa.
The rapid increase in antimicrobial resistance driven by the widespread use, abuse, and misuse of antibiotics constitutes one of China’s most challenging healthcare problems. In particular, nosocomial infections caused by multidrug-resistant organisms such as methicillin-resistant Staphylococcus aureus (MRSA), carbapenem-resistant Acinetobacter baumannii (CRAB), and carbapenem-resistant Enterobacterales (CRE), which exhibit resistance to most available antibiotics, lead to high mortality and enormous economic and human costs. Here, we summarize the current patterns of the antimicrobial resistance of nosocomial infections in China and address possible interventions to combat antimicrobial resistance.
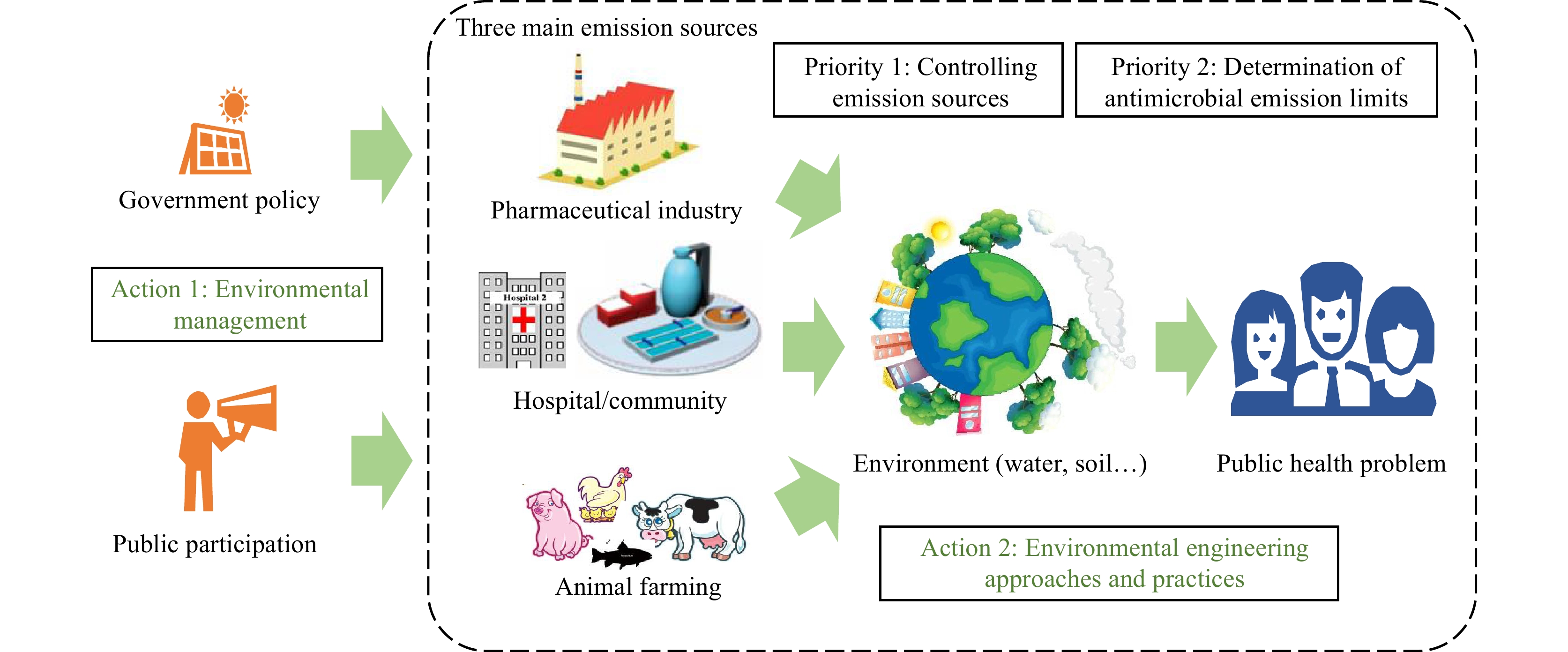
Antimicrobial resistance (AMR) is a globally recognized crisis with meaningful engagement across humans, animals, and the environment as in the One Health approach. The environment is the potential source, reservoir, and transmission route of AMR, and it plays a key role in AMR development from the One Health perspective. Animal farming, hospitals, and the pharmaceutical industry are identified as the main emission sources in the environment. Minimizing emissions and determining antimicrobial emission limits are priorities in the containment of environmental AMR development. From the perspectives of environmental management and environmental engineering, some important actions to minimize risks of AMR development are summarized, including the recent progress in enhanced hydrolysis pre-treatment technology to control the development of antibiotic resistance genes (ARGs) during biological wastewater treatment. It is desirable to establish a holistic framework to coordinate international actions on the containment of environmental AMR development. To establish a community with a shared future for humanity, China should and could play an important role in international cooperation to cope with AMR challenges.
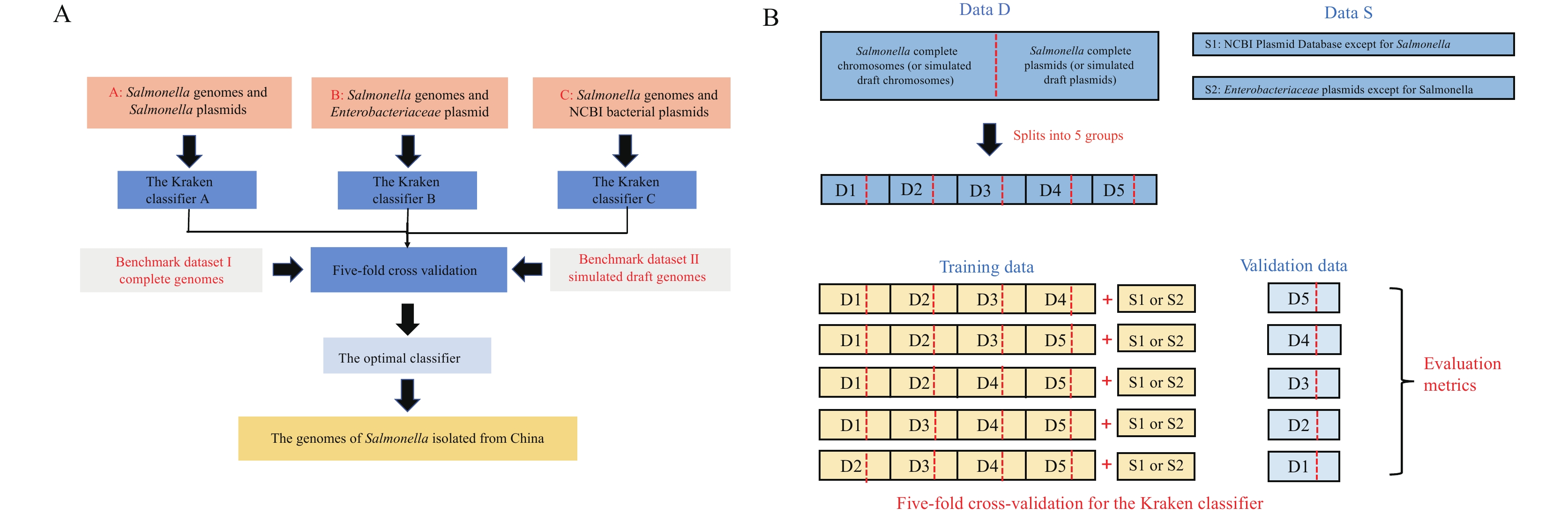
Salmonella is a key intestinal pathogen of foodborne disease, and the plasmids in Salmonella are related to many biological characteristics, including virulence and drug resistance. A large number of plasmid contigs have been sequenced in bacterial draft genomes, however, these are often difficult to distinguish from chromosomal contigs.
In this study, three different customized Kraken databases were used to build three different Kraken classifiers. Complete genome benchmark datasets and simulated draft genome benchmark datasets were constructed. Five-fold cross-validation was used to evaluate the performance of the three different Kraken classifiers by two benchmark datasets.
The predictive performance of the classifier based on all National Center for Biotechnology Information plasmids and Salmonella complete genomes was optimal. This optimal Kraken classifier was performed with Salmonella isolated in China. The plasmid carrying rate of Salmonella in China is 91.01%, and it was found that the Kraken classifier could find more plasmid contigs and antibiotic resistance genes (ARGs) than results derived from a plasmid replicon-based method (PlasmidFinder). Moreover, it was found that in the strains carrying ARGs, plasmids carried more ARGs [three, 95% confidence interval (CI): 1–14] than chromosomes (one, 95% CI: 1–7).
We found building a high-quality customized database as a Kraken classifier to be ideal for the prediction of Salmonella plasmid sequences from bacterial draft genomes. In the future, the Kraken classifier established in this study will play a significant role in ARG monitoring.



 Subscribe for E-mail Alerts
Subscribe for E-mail Alerts CCDC Weekly RSS Feed
CCDC Weekly RSS Feed