-
We read the study conducted by Cui et al. published in China CDC Weekly with great interest, and would like to make comment on this matter (1). The report identified 31 Salmonella enterica serovar I 1,4,[5],12:i:- (S. I 1,4,[5],12:i:-) sequence type 8333 (ST8333) genomes by the end of 2023 in the National Molecular Tracing Network for Foodborne Disease Surveillance database. The overall percentage of isolates of S. I 1,4,[5],12:i:- ST8333 remained low in their active monitoring between 2017 and 2023, and observed ST8333 in 2017.
In fact, ST8333 was first assigned in the EnteroBase database (https://enterobase.warwick.ac.uk/) based on 7 housekeeping genes (including aroC: 10, dnaN: 19, hemD: 1050, hisD: 9, sucA: 9, purE: 5, and thrA: 2) on January 15, 2021 (Figure 1A) (2). The first S. I 1,4,[5],12:i:- ST8333 strain was isolated from a case of sporadic diarrhea (a 1-year-old girl) in the Xinjiang Uygur Autonomous Region, China, on August 24, 2017 (2). In our previous study, we built an open-access Chinese Local Salmonella Genome Database version 2 (CLSGDB v2, https://nmdc.cn/clsgdbv2), which consisted of 7,997 high-quality genomes with 164 serovars and 295 STs, including two S. I 1,4,[5],12:i:- ST8333 genomes (3). In addition to our previous report (2), another S. I 1,4,[5],12:i:- ST8333 strain was isolated from frozen raw ground pork in Hanzhong City, Shaanxi Province, China in August 2019 (3). Cui et al. reported 31 ST8333 genomes from 4 provincial-level administrative divisions (PLADs) in China, thereby increasing the knowledge of the prevalence and distribution of ST8333 (1). Based on our recent sequencing data and mining of publicly available databases as of June 12, 2025 (including NCBI, Enterobase, and CLSGDBv2) (3), we found that S. I 1,4,[5],12:i:- ST8333 was isolated from a 1-year-old boy in Xinjiang as early as July 25, 2015. Therefore, we depicted the timeline of ST8333 for its existence and discovery to help us better understand the spread of this important ST (Figure 1A). According to the date of isolation, at least six S. I 1,4,[5],12:i:- ST8333 isolates had existed in Xinjiang before 2017. Three of these were isolated from children under 5 years of age.
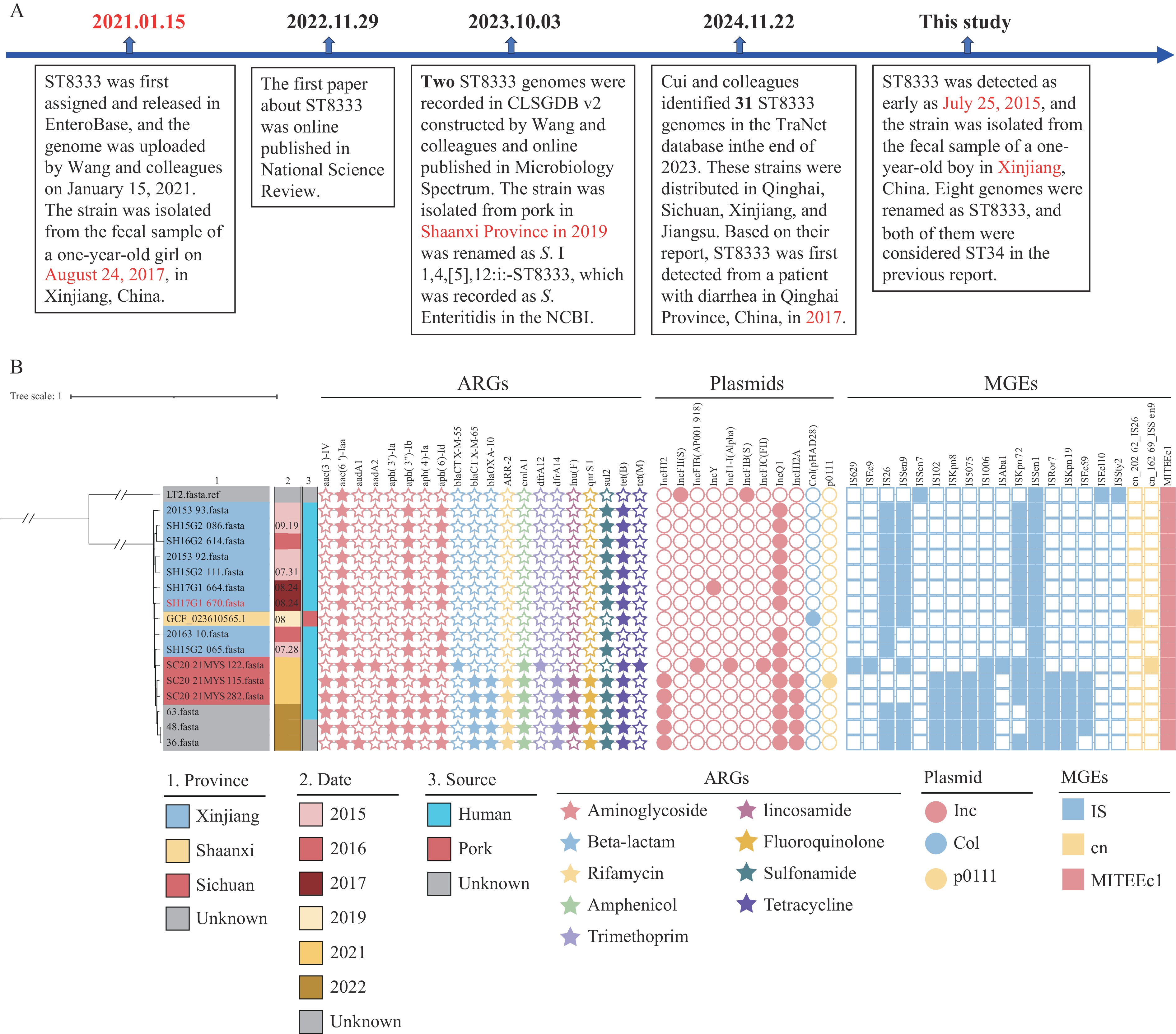 Figure 1.
Figure 1.Discovery timeline and phylogenetic analysis of S. I 1,4,[5],12:i:- ST8333. (A) Timeline of ST8333 for its existence and discovery; (B) Phylogenetic analysis of S. I 1,4,[5],12:i:- ST8333 genomes.
Note: Four strains, including DS23IMWYN_873, SH17G1670, SH15G2065, and SH15G2086, were isolated from children under 5 years old; SH15G2111 and SH17G1664 were isolated from 17 and 35 years old, respectively; the others were unknown. The core genome phylogeny and multi-alignments were performed using the Parsnp v2.1.0 (https://github.com/marbl/parsnp) and visualized using the Interactive Tree of Life (iTOL version 7.0, https://itol.embl.de/). 2015393: SAMN35670174; 2015392: SAMN35670173; SH15G2065: SAMN35670843; SH15G2086: SAMN35670847; SH15G2111: SAMN35670848; 2016310: SAMN35670191; SH16G2614: SAMN35680157; SH17G1664: SAMN35680386; SH17G1670: SAL_FB7519AA (SAMN21890000); 36: SAL_PB8236AA; 48: SAL_PB8237AA; 63: SAL_OB7431AA; SC2021MYS115: SAL_NB5716AA; SC2021MYS122: SAL_NB5708AA: SC2021MYS282: SAL_NB5698AA. DS23IMWYN237_870: SAMN48529803. For (A), the timeline in the diagram briefly describes the significant stories at each time point. For (B), location, collection date, isolation source, ARGs, plasmid replicons, MGEs, and SPIs are indicated with different colors and shapes, respectively.
Abbreviation: ARGs=antibiotic resistance genes; MGEs=mobile genetic elements; SPIs=Salmonella Pathogenic Islands.
Based on the phylogenetic analysis of 31 S. I 1,4,[5],12:i:- ST8333 genomes, Cui et al. concluded that the ST8333 strain originated in Qinghai Province in 2017 and then spread to Xinjiang, Sichuan, and Jiangsu PLADs, China (1). However, because of the limited genomic data used, their conclusions may exist certain limitations in terms of accuracy. Our analysis indicated that endemic transmission (the presence of a like-outbreak) occurred in Xinjiang in July 2015, and then spread to other PLADs, such as Sichuan and Shaanxi (Figure 1B). These findings suggest that the location where the ST8333 strain was first isolated does not necessarily represent the location where the ST first appeared. In addition, due to the limited release of the genomic data (1), we were unable to conduct comparative genomic analyses to obtain a better evolutionary analysis. Therefore, we recommend further data sharing as soon as possible.
Further in silico analysis showed a total of 124 acquired antibiotic resistance genes (ARGs); 6 strains carried third-generation cephalosporin resistance genes, including blaCTX-M-65 and blaCTX-M-55, and 5 strains carried the fluoroquinolone resistance gene qnrS1. Very few ARGs, plasmid replicons, and mobile genetic elements (MGEs) were identified in earlier isolates (i.e., 2015) compared to genomes isolated in recent years (Figure 1B). The co-existence of several ARGs (qnrS1, blaOXA-10, blaCTX-M-65, cmlA1, lun(F), dfrA14, and ARR-2), IncHI2 replicon, and MGEs (IS102, IS5075, IS1006, ISRor7, ISKpn8, and ISKpn19) in strains isolated in 2021 and 2022 was observed (Figure 1B).
Recently, S. I 1,4,[5],12:i:- has become the most prevalent cause of human infections in China, and ST34 is the predominant ST among human infection cases in China (1,3). Comparative genomics confirmed that ST8333 evolved from S. I 1,4,[5],12:i:-ST34 (2), which is prone to multidrug resistance (MDR). Based on limited data, the number of infection cases caused by ST8333 is increasing, and the ST8333 strain has become increasingly severe owing to the acquisition of ARGs. It remains uncertain whether ST8333 will replace ST34 as the primary sequence type clone that causes human infections worldwide. These findings highlight the need for global public health attention and urgent genomic monitoring.
The emergence and increasing trends of antimicrobial resistance (AMR) worldwide are among the top 10 public health threats (4). S. enterica is listed on the 2024 World Health Organization Bacterial Priority Pathogens List (5–6), and the increasing trends of AMR in S. enterica are of great public health concern (2–3,7). With recent advances in large data-empowered artificial intelligence (AI) and machine learning, WGS has become critical for tracking the rapid spatiotemporal evolution of AMR in S. enterica (2–3,7–11). These findings underscore the importance of high-quality, open-access regional and international genomic databases for fighting against AMR, ARGs, and informing outbreak response (11–12). Therefore, more publicly available data on S. I 1,4,[5],12:i:- and S. Typhimurium will help to better evaluate the spread situation in real-time and enable the scientific community to formulate and adjust prevention and control policies in a prompt manner. Failure to fill these gaps may lead to underestimation of the risk of transmission of the MDR ST8333 clone and the emergence of new variants in S. I 1,4,[5],12:i:-. Considering the positive correlation between the gross output of meat, population density, annual mean temperature, and AMR in Salmonella (3,7), as well as the emergence of MDR ST8333 in meat (pork), we call for more S. I 1,4,[5],12:i:- and S. Typhimurium strains from meat and animal sources for WGS and data sharing in the open-access Salmonella genome database.
HTML
-
The Veterinary Big Data and Bioinformatics Center, Henan Agricultural University, for their support and help.
| Citation: |



 Download:
Download:




