-
Brucellosis remains a significant global zoonosis with profound negative implications for human health and economic systems worldwide (1). The disease is caused by Gram-negative, facultative intracellular bacteria belonging to the genus Brucella (2). Currently, 13 recognized Brucella species exist, with B. melitensis, B. abortus, and B. suis representing the predominant human pathogens (3). Human infection with Brucella spp. typically occurs through consumption of contaminated animal products or direct contact with infected animals (4).
In August 2024, a 71-year-old male farmer from Baodi District, Tianjin Municipality, China presented with persistent fever, excessive sweating, muscle and joint pain, and fatigue. The patient reported regularly entering a sheep pen without protective equipment approximately every 7 days, though he had no history of travel to known brucellosis-endemic regions. A Brucella strain, designated “BD002,” was isolated from the patient, confirming the diagnosis of brucellosis. Epidemiological investigation suggested that the sheep in the patient’s care were the likely source of infection.
For isolation of the Brucella strain, 500 μL of blood sample was inoculated onto a blood agar plate and incubated at 37 °C with 5% carbon dioxide for 72 hours, yielding off-white single colonies. The bacterial suspension was subsequently prepared and inactivated for DNA extraction and polymerase chain reaction (PCR) analysis. The brucella cell surface protein 31 polymerase chain reaction (BCSP31-PCR) test for genus identification revealed the characteristic 224-bp DNA band, while the abortus, melitensis, ovis, and suis polymerase chain reaction (AMOS-PCR) species-identification test yielded no detectable DNA bands (5). Notably, AMOS-PCR can identify B. abortus biovars 1, 2, and 4, B. melitensis, and B. ovis, but does not encompass all biovars of Brucella spp.
To further characterize strain BD002, we conducted both multi-locus sequence typing (MLST) and next-generation sequencing (NGS) through GENEWIZ Co. and BGI-Shenzhen, China, respectively. For MLST, 9 Brucella housekeeping genes were amplified and subjected to Sanger sequencing, which identified BD002 as sequence type 2 according to the PubMLST database. NGS analysis generated 23 contigs using SPAdes v3.7, with a total genome length of 3,252,744 base pairs (6). The assembled genomic sequences have been deposited in the National Genomics Data Center (Accession No. GWHFJIX00000000.1) (7). We conducted phylogenetic analysis using the BD002 genome alongside 386 B. abortus genomic sequences from the NCBI Genome database, employing Snippy v4.6 and RAxML-ng v1.2.2 with strain 544 (Accession No. GCA_000369945.1) as the reference sequence for core-genome single-nucleotide polymorphism analysis (8). The resulting maximum likelihood phylogenetic tree was visualized using R v4.3.1 and ggtree v3.10.1 (9). Phylogenetic analysis revealed 4 distinct clades, with BD002 clustering within Clade B (Figure 1A). Notably, BD002 formed a sub-clade with strains from other Chinese provincial-level administrative divisions, including Inner Mongolia, Jilin, Heilongjiang, Beijing, Hebei, and Shanxi, as well as strains from Russia, Georgia, Mongolia, and other countries (Figure 1B).
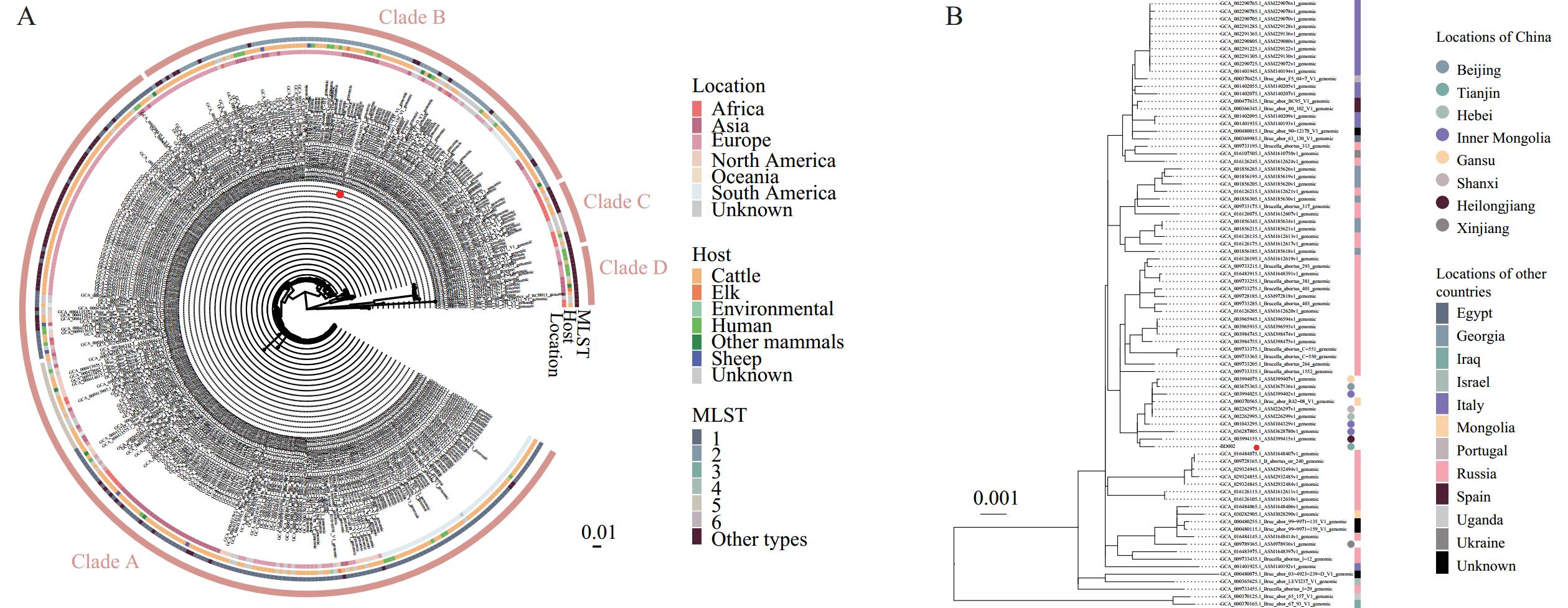 Figure 1.
Figure 1.The ML phylogenetic tree of B. abortus genomes based on cg-SNP. (A) The ML phylogenetic tree was constructed using the genomic sequence of strain BD002 integrated with 386 additional B. abortus genomic sequences and associated metadata obtained from the NCBI genome database. (B) A detailed view of the subclade within clade B containing strain BD002 is presented, with countries of strain origin indicated by differently colored circles. The BD002 strain is specifically highlighted with a red circle.
Note: For (A), the analysis revealed four distinct worldwide clades of B. abortus strains, highlighted in orange. The continental origin, host species, and sequence types of the strains are represented by differently colored squares. Abbreviation: ML=maximum likelihood; cg-SNP=core-genome single-nucleotide polymorphism.Human brucellosis in Tianjin affects approximately 200 individuals annually, with B. melitensis identified as the predominant causative agent. This study documents the first isolation of B. abortus from a human case in Tianjin, indicating the circulation of multiple Brucella species within the region.
Compared with traditional AMOS-PCR methodology, MLST and NGS techniques provide superior accuracy in Brucella spp. identification and offer substantially higher resolution for phylogenetic analysis. These advanced molecular approaches should be fully integrated into routine etiological surveillance of human brucellosis.
Additionally, particular attention should be directed toward livestock handlers who neglect to appropriate personal protective equipment and fail to implement adequate disinfection and hygiene practices following occupational exposure (10).
HTML
| Citation: |



 Download:
Download:




