-
Wastewater-based epidemiology (WBE) has emerged as a pivotal and cost-effective public health tool, offering a swift and efficient means to evaluate the prevalence and dissemination of pathogens within a community through wastewater analysis (1). This method supports a comprehensive approach called One Sample, Multiple Analyses (OSMA) (2), allowing simultaneous monitoring of various health indicators, including antibiotic resistance genes, drug-resistant bacteria, pathogens, and chemical contaminants. This enhances traditional surveillance techniques. WBE’s non-intrusive nature augments clinical surveillance by detecting pathogens in wastewater, which helps provide early warnings of disease outbreaks and track infection trends (3-4).
The utility of WBE was especially evident during the COVID-19 pandemic, where it effectively traced SARS-CoV-2 transmission, demonstrating its value in infectious disease surveillance (4-5). The monitoring of enteric pathogens including Salmonella, Shigella, and Vibrio species in wastewater has provided crucial epidemiological insights into foodborne and waterborne diseases. These pathogens, responsible for conditions like gastroenteritis, dysentery, and cholera, are consistently detected in wastewater systems (6–8). Surveillance of these pathogens is vital for understanding disease patterns, assessing intervention effectiveness, identifying contamination sources, quantifying disease burden, and guiding targeted surveillance efforts.
Digital polymerase chain reaction (dPCR), a third-generation PCR technology, provides exceptional accuracy and sensitivity for absolute quantification. This study establishes a quadruplex dPCR method for the simultaneous detection of Salmonella, Shigella, V. parahaemolyticus, and V. cholerae in wastewater samples. By monitoring bacterial levels in wastewater with dPCR, this approach serves as an early warning system, enabling timely identification of outbreak risks and prompt implementation of preventive measures.
-
A total of 60 wastewater samples were collected from piped wastewater networks serving large residential communities, subway stations, educational institutions, and hospitals across 4 districts of Beijing Municipality (Chaoyang, Haidian, Daxing, and Fengtai), China. The collected samples were categorized as follows: 27 from residential communities, 18 from healthcare institutions, and 15 from other locations. All samples were collected in November 2024.
-
Fresh bacterial colonies of the four positive control strains were washed three times with phosphate-buffered saline (PBS, pH 7.4) and subjected to serial twofold dilutions in PBS. For each dilution, 1 mL was spiked into 100 mL of pre-screened wastewater that had tested negative for all four target pathogens.
-
The positive control strains utilized for method development were V. cholerae strain N16961, Salmonella enterica serovar Typhimurium strain LT2 (9), S. flexneri strain SH1, and V. parahaemolyticus strain ICDC-VP1329. The specificity of the dPCR assay was validated using a comprehensive panel of 90 bacterial strains, comprising 51 target strains and 39 non-target strains. The panel included 16 Salmonella strains (5 S. Enteritidis, 5 S. typhi, and 6 S. Typhimurium); 9 Shigella strains (2 S. flexneri and 7 S. sonnei); 11 V. parahaemolyticus; 13 V. cholerae (7 O1 serogroup and 6 O139 serogroup, with 12 ctxA-positive strains); 2 Enteroinvasive Escherichia coli (EIEC); 20 other pathogenic E. coli (excluding EIEC); and various other species including 1 V. alginolyticus, 3 V. fluvialis, 2 V. mimicus, 2 V. vulnificus, 1 Aeromonas hydrophila, 3 Plesiomonas shigelloides, 1 Yersinia enterocolitica, 2 Clostridium perfringens, 1 Edwardsiella tarda, 1 Listeria monocytogenes, 1 Citrobacter sp., and 1 Clostridium difficile. All strains were maintained in laboratory collection.
-
Target gene sequences were selected based on comprehensive sequence alignments from the NCBI NR database (https://www.ncbi.nlm.nih.gov/nucleotide/). All primers and probes were designed using Beacon Designer V8.20 and synthesized by Sangon Biotech (Shanghai, China).
The primer and probe sequences (5’ to 3’) for pathogen detection are as follows: for the invA gene of Salmonella, the forward primer is CCGCCAAACCTAAAACCAG, the reverse primer is GGCTCTTCGGCACAAGTA, and the probe is FAM-CGCCAATCAGTCCYAACGACGACCCTT-BHQ1; for the ipaH gene of Shigella, the forward primer is GCAGAGAAACTTCAGCTCTC, the reverse primer is CAGTGCGGAGGTCATTTG, and the probe is HEX-TCACTCCCGACACGCCATAGAAACGC-BHQ1; for the tlh gene of V. parahaemolyticus, the forward primer is CGAACGAGAACGCAGACATTA, the reverse primer is GCAACCACTTTGTTGATTTGATCT, and the probe is ROX-TTCTTCGCCGCTGACAATCGCTTCTCA-BHQ2; and for the ctxA gene of Vibrio cholerae, the forward primer is AGGGGCTACAGAGATAGATATTACA, the reverse primer is GCGGTGCATGATGAATCCA, and the probe is Cy5-ACCTGCCAATCCATAACCATCTGCTGC-BHQ3.
-
The quadruplex dPCR analysis was performed using the multiplex QIAcuity Digital PCR system (QIAGEN, Hilden, Germany). Reactions were conducted using the QIAcuity Probe PCR Kit (QIAGEN) according to manufacturer specifications. Each 40 µL reaction mixture contained 10 µL of 4× dPCR™ Supermix for Probes (QIAGEN), 10 µL of template DNA, 800 nM of each primer, 400 nM of each probe, and 6.25 units/mL EcoRI-HF® (New England Biolabs, United Kingdom). The reaction mixture was distributed into a 24-well 26k QIAcuity Nanoplate (QIAGEN), sealed, and analyzed using the QIAcuity dPCR instrument (QIAGEN). Each sample was analyzed in triplicate, with a no-template control (NTC) serving as the negative control. The thermal cycling protocol consisted of initial denaturation at 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing/extension at 58 °C for 30 s. Positive results were determined by the presence of positive droplets in the dPCR assay.
-
The specificity of primers and probes underwent comprehensive evaluation using both the BLASTn algorithm (within the non-redundant nucleotide database) and Primer-BLAST (within the non-redundant database restricted to Enterobacterales) (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Subsequent validation was performed against genomic DNA extracted from 90 bacterial strains, comprising 51 target strains and 39 non-target strains as detailed above.
-
The analytical sensitivity of the assay was determined using artificially spiked wastewater samples. Bacterial concentrations in each dilution were quantified by colony-forming unit (CFU) counting on LB agar plates. The spiked wastewater samples underwent centrifugation at 10,000 g for 30 minutes, and DNA was extracted from the resulting pellet using a DNeasy® PowerSoil Kit (QIAGEN, USA) according to the manufacturer’s protocol. The extracted DNA was resuspended in 100 μL of nuclease-free water, with 10 μL used as template for dPCR detection. The entire process, from spiked sample preparation through dPCR detection, was performed in triplicate. The LOD was defined as the lowest CFU concentration in wastewater that yielded positive results across all replicates(10). Process repeatability was assessed using the coefficient of variation (CV) of the measured copy number per reaction.
-
The dPCR data were analyzed using the QIAcuity Software Suite (version 2.5.0.1; Qiagen, Germany). A database was established using Microsoft Excel 2010. Statistical analysis and visualization were conducted using R software (version 4.4.2, R Core Team, Vienna, Austria). Differences in target gene concentrations in nucleic acid extracted from 100 mL wastewater samples across different sampling sites were assessed using the Wilcoxon test, implemented with the “wilcox_test” function from the “rstatix” package. Whisker plots were created using the “ggplot2” package.
-
Following primer and probe design, the amplification efficiency of each target gene using serial dilutions of positive control strain was initially evaluated DNA via real-time PCR. The analysis demonstrated optimal amplification efficiency (90%–110%) for each target gene in both singleplex (single target gene with corresponding primers and probe) and quadruplex (all four target genes with corresponding primers and probes) systems. After confirming amplification efficiency, the quadruplex dPCR reaction conditions were optimized. Through systematic evaluation of annealing/extension temperatures between 58°C and 62°C, 58°C provided optimal discrimination between positive and negative partitions. Based on these results, 58°C was the optimal annealing and extension temperature for the quadruplex dPCR assay (Figure 1).
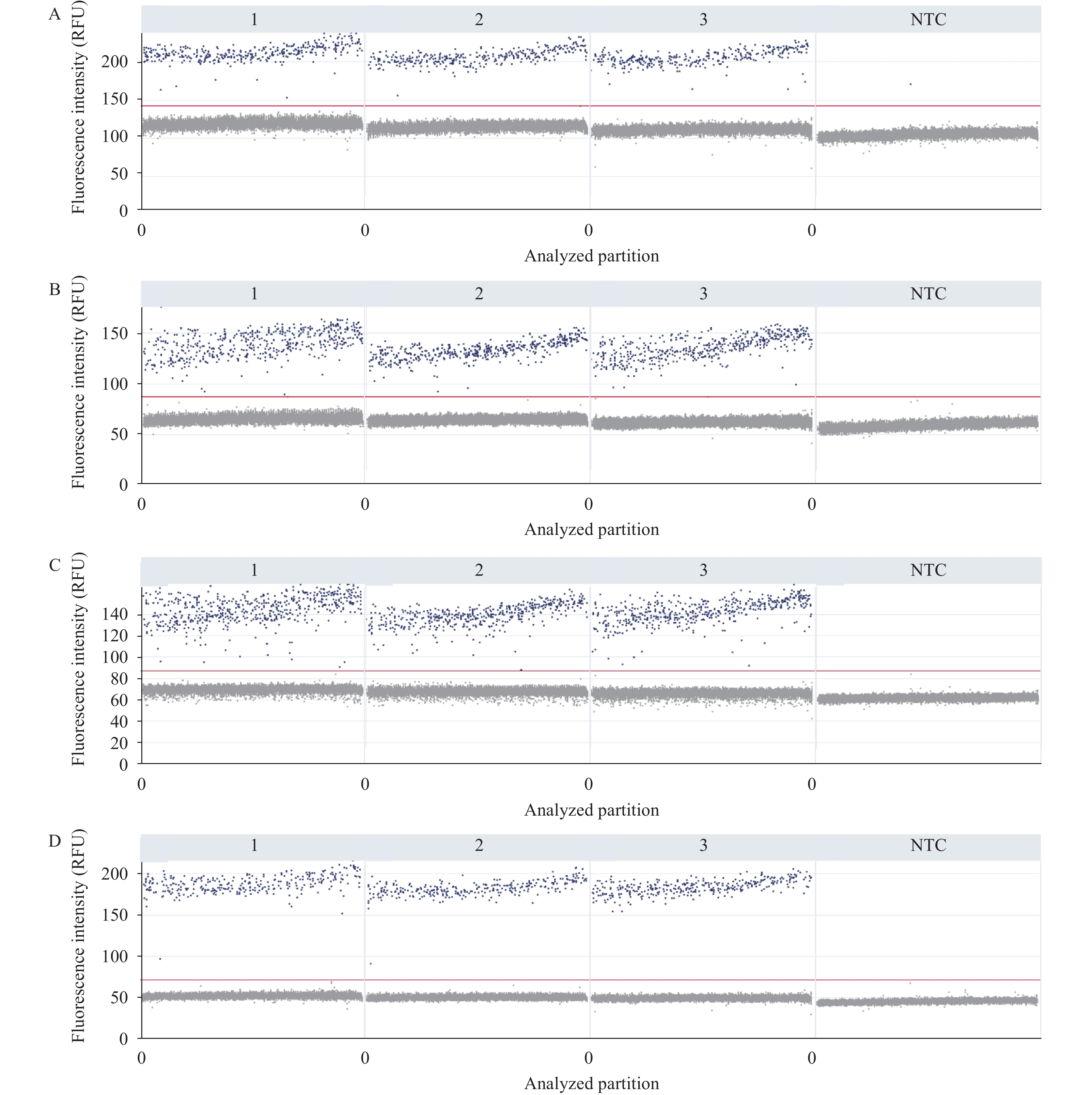 Figure 1.
Figure 1.The fluorescence intensity of the target gene by digital PCR. (A) the ipaH gene of Shigella SH1; (B) the invA gene of Salmonella LT2; (C) the cholera toxin gene ctxA of Vibrio cholerae N16961; (D) the tlh gene of Vibrio parahaemolyticus ICDC-VP1329. 1, 2, and 3: Three replicates.
Note: The red horizontal lines denote the fluorescence threshold. Partitions represented by blue dots above this threshold line are interpreted as positive, while those represented by grey dots positioned below the threshold line are classified as negative.
Abbreviation: NTC=no-template control; PCR=polymerase chain reaction; RFU=relative fluorescence unit.
-
In silico specificity analysis confirmed high target specificity for all primers and probes across the four target genes. However, two important considerations emerged: the ipaH gene is present in both Shigella and EIEC, and while the ctxA gene is predominantly found in V. cholerae, it can occasionally occur in other species such as V. mimicus. Subsequent experimental validation using 90 bacterial strains (51 target and 39 non-target) demonstrated robust analytical specificity of the quadruplex dPCR assay. The assay showed complete concordance with expected results for target organisms, with no observed cross-reactivity or false-positive results across the tested strain panel.
Using artificially spiked wastewater samples, the following LOD values were established for the quadruplex dPCR assay: 390 CFU/100 mL for Salmonella, 11 CFU/100 mL for Shigella, 660 CFU/100 mL for V. cholerae, and 640 CFU/100 mL for V. parahaemolyticus (Table 1).
Bacterial strains Expected CFU/mL of wastewater sample Mean±SD
(cp/µL DNA)CV (%) LOD (CFU/100 mL wastewater sample) Shigella flexneri SH1 0.22 3.13±0.31 9.78 0.11 1.31±0.36 27.49 11 0.05 1.16*±0.33 28.65 Salmonella enterica serovar Typhimurium strain LT2 7.80 9.65±1.01 30.5 3.90 3.33±1.02 30.5 390 1.95 0.95*±0.33 35.22 Vibrio cholerae N16961 33.00 37.48±15.81 42.17 6.60 2.23±1.39 62.30 660 3.30 0.15*±0.13 87.12 Vibrio parahaemolyticus ICDC-VP1329 12.80 11.45±1.11 9.7 6.40 4.56±0.40 8.83 640 3.20 1.38*±0.54 38.79 Notes: Mean, average copy values (n=3) in a final reaction volume of 40 μL. CV between replicates (n=3), CV=SD/Mean ×100%. Expected CFU/mL of wastewater sample: the bacterial concentration calculated by colony counting results.
Abbreviation: CV=coefficient of variation; SD=standard deviation; LOD=limit of detection; CFU=colony-forming unit.
* In three replicates, a negative well appeared.Table 1. Results of dPCR for the detection of artificially spiked wastewater samples.
-
The quadruplex dPCR assay was employed to analyze four bacterial pathogens across 60 municipal wastewater samples collected from the pipe network. Analysis revealed differential detection rates among the target pathogens: Shigella exhibited the highest prevalence at 78.3% (47/60), followed by Salmonella at 46.7% (28/60), and V. parahaemolyticus at 33.3% (20/60) (Table 2). No V. cholerae was detected in any of the samples. Quantitative analysis demonstrated pathogen-specific concentration ranges: Shigella showed the highest concentrations at 100.9–14,560 copies/100 mL, followed by Salmonella at 86.5–7,329 copies/100 mL, and V. parahaemolyticus at 84.5–865.7 copies/100 mL (Figure 2).
Type of Place Shigella spp. Salmonella spp. Vibrio parahemolyticus Vibrio cholerae POS
(%)POS/SUM POS
(%)POS/SUM POS
(%)POS/SUM POS (%) POS/SUM Community 88.9 24/27 63.0 17/27 40.7 11/27 0.0 0/27 Hospital 53.3 8/15 13.3 2/15 13.3 2/15 0.0 0/15 Other 83.3 15/18 50.0 9/18 38.9 7/18 0.0 0/18 Total 78.3 47/60 46.7 28/60 33.3 20/60 0.0 0/60 Notes: Positive well: a well is considered positive when the number of positive droplets is greater than or equal to 3. Positive sample: a sample is determined to be positive when two or more of its three replicate wells are positive.
Abbreviation: POS=number of positive samples. SUM=the sum of positive and negative samples.Table 2. Comparison of the quadruplex dPCR for detecting 60 wastewater samples of different types (community n=27, hospital n=15, other n=18) collected from Beijing Municipality, China.
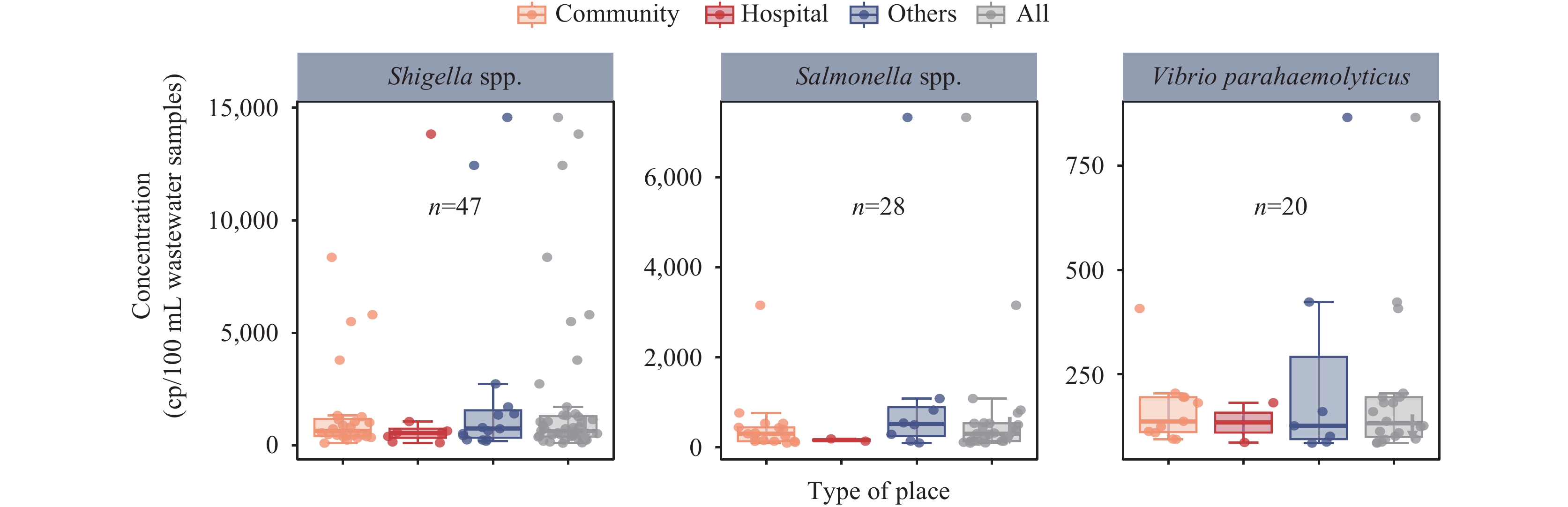 Figure 2.
Figure 2.Whisker plots of the concentration of the target genes in the nucleic acid extracted from 100 mL wastewater samples from different sampling sites measured by quadruplex dPCR. Upper and lower fences display the 5th and 95th percentiles. The boxes display the interquartile range and median.
Abbreviation: dPCR=digital polymerase chain reaction. -
Current approaches for pathogen detection in wastewater encompass traditional culture-based methods, molecular techniques such as PCR, and next-generation sequencing (11). However, these methodologies exhibit significant limitations in sensitivity, specificity, and their capacity to detect low pathogen concentrations in complex wastewater matrices. dPCR has emerged as a transformative technology for wastewater surveillance, offering superior sensitivity and quantitative precision for pathogen detection (12). Its capacity for absolute quantification without calibration curves renders dPCR particularly advantageous for wastewater analysis, especially given its robust performance in the presence of PCR inhibitors commonly encountered in complex environmental matrices (12-13).
Previous studies have demonstrated the efficacy of multiplex dPCR for bacterial detection. A duplex dPCR assay developed for Shigella and Salmonella detection in fecal samples achieved detection limits of 12.3 copies/r and 23.7 copies/r, with corresponding CFU/mL values of 550 and 10,000(14), respectively. In this study, the detection limits were 13.1 copies/r and 33.3 copies/r. Comparatively, this method demonstrates enhanced sensitivity, with detection limits of 11 and 390 CFU/100 mL for these bacteria in wastewater matrices. These performance differences may be attributed to variations in sample type and processing procedures. While another triplex dPCR assay reported a detection limit of 0.23 copies/µL for Salmonella nucleic acid in food samples, our method achieved 3.33 copies/µL in wastewater matrices. This sensitivity variation likely reflects differences in both methodological approaches and matrix-specific bacterial recovery rates. The disparity underscores the critical importance of optimizing wastewater sample pretreatment protocols for real-world applications. In this analysis of environmental samples, the elevated detection rates for Shigella and Salmonella, with concentrations predominantly ranging from 394.55 to 1300 and 131.9 to 527.3 CFU/100 mL, respectively, indicate potential public health concerns that warrant enhanced surveillance measures. This sensitivity variation is likely attributable to differences in methodological approaches, as well as potential variations in DNA extraction and PCR amplification efficiency, which may be influenced by matrix differences. These discrepancies highlight the critical need for standardized wastewater sample pretreatment methods in surveillance systems, particularly when comparing data across different sentinel sites. Since the ipaH gene is present in both Shigella and EIEC, and the ctxA gene can occasionally be found in other species, for positive detection of Shigella and Vibrio cholerae, it is recommended to isolate and culture the strains before performing biochemical identification. Alternatively, detection can focus on specific genes of EIEC and vibroid-like bacteria.
This study presents the first application of multiplex dPCR for simultaneous monitoring of these four pathogens in wastewater environments. The developed quadruplex dPCR method enhances detection throughput and facilitates large-scale wastewater monitoring programs. The method’s capacity to detect pathogens at low concentrations enables earlier identification of potential disease outbreaks.
Despite these advantages, several challenges and limitations remain in implementing multiplex dPCR for wastewater monitoring. First, the technology has not yet achieved full process automation and still relies on operator technical proficiency, potentially affecting result accuracy. Second, samples with extremely low concentrations often exhibit high CV values across repeated measurements, with discrepancies between detected values and actual copy numbers — a phenomenon which was also encountered in this experiment.
Therefore, to enhance result reliability and accuracy in practical applications, it is recommended to perform at least three replicate measurements and calculate the average. Furthermore, while dPCR assays remain relatively costly and the complexity of analysis may pose barriers for laboratories with limited resources, potentially hindering widespread application in public health monitoring, exploring collaborations with external laboratories to share equipment or providing simplified data analysis software could help lower entry barriers. Due to the nascent stage of wastewater monitoring in our country and the absence of standardized protocols for multiplex dPCR in wastewater surveillance, result variability across different laboratories has hindered cross-study comparability. Therefore, establishing national standards promptly is essential to unify workflows and practices among laboratories.
-
The Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences and the Chinese Urban Wastewater Surveillance System for their valuable support.
-
This study did not involve human or animal subjects, thus, no ethical statement was required.
HTML
Wastewater Samples
Artificially Spiked Wastewater
Bacterial Strains
Primers and Probes
Quadruplex dPCR Assay
Analytical Specificity
Limit of Detection (LOD)
Statistical Analysis
Establishment and Optimization of the Quadruplex dPCR Assay for Four Bacterial Pathogens
Analytical Specificity and Sensitivity
Pathogen Detection and Quantification in Municipal Wastewater Samples Using Quadruplex dPCR Assay
| Citation: |



 Download:
Download:




