HTML
-
Introduction: This study aims to analyze the clinical characteristics and pathogenic features of Chlamydia pneumoniae pneumonia, providing a scientific basis for the diagnosis and treatment of C. pneumoniae infections.
Methods: Clinical data from four patients diagnosed with C. pneumoniae pneumonia in Lishui City, Zhejiang Province, between April and May 2024, were collected to analyze clinical manifestations and pathogenic findings. Multi-locus sequence typing (MLST) analysis of the pathogen was conducted using seven housekeeping genes.
Results: All patients exhibited decreased levels of retinol-binding protein and prealbumin, findings not previously reported in earlier studies. Additionally, co-infections were identified in two cases. Analysis of the 16S rRNA and ompA gene sequences indicated a homology of 98% to 100% with known C. pneumoniae strains. To further characterize these strains, sequencing of the seven housekeeping genes confirmed that all cases were infected with the ST16 genotype.
Conclusions: C. pneumoniae infections in Lishui City are predominantly caused by the ST16 genotype, highlighting the need for enhanced research into these infections. The decrease in retinol-binding protein and prealbumin levels may serve as auxiliary diagnostic biomarkers in clinical practice. Next-generation sequencing methods demonstrate significant potential for pathogen identification, particularly in diagnosing co-infections.
-
Humans are the only known reservoir of Chlamydia pneumoniae (C. pneumoniae), a gram-negative, obligate intracellular bacterium. This pathogen represents a common etiological agent of community-acquired pneumonia, with diagnosis and treatment frequently delayed due to its presentation with atypical clinical manifestations. Beyond respiratory infections, emerging evidence suggests associations between C. pneumoniae and various pathological conditions, including asthma, atherosclerosis, and Alzheimer disease (1).
In China, neither C. pneumoniae pneumonia nor C. psittaci pneumonia are classified as notifiable infectious diseases, and these pathogens are not routinely included in standard clinical diagnostic panels for respiratory infections. Consequently, the epidemiological prevalence and genotypic distribution of chlamydial infections remain inadequately characterized. To date, only a single serovar of C. pneumoniae has been identified. Previously, our research team investigated the prevalence of C. psittaci infections in Lishui, revealing the presence of a novel C. psittaci strain responsible for local infections (2). In the current study, to elucidate the prevalence and genotypic characteristics of C. pneumoniae in Lishui, we conducted comprehensive clinical and laboratory investigations of four cases of severe community-acquired pneumonia.
In this study, an analysis was conducted for consecutive cases of C. pneumoniae infection in patients admitted to Lishui People’s Hospital of Zhejiang Province between April 23 and May 14, 2024. Nested polymerase chain reaction targeting Chlamydia-specific genes of 16s rRNA and outer membrane protein A (ompA) were amplified. To further characterize the C. pneumoniae isolates through multi-locus sequence typing (MLST), we employed nested polymerase chain reaction methods to sequence seven housekeeping genes (enoA, fumC, gatA, gidA, hemN, hflX, and oppA) (3). Targeted next-generation sequencing (tNGS) was utilized for pathogen diagnosis in patients 2 and 3. Additionally, we attempted to obtain the complete genome sequence of C. pneumoniae using the NGS-based hybrid capture method.
The study cohort comprised three females and one male, with a median age of 22 years (range: 14–36 years). Medical records indicated that three patients presented with high fever. All four patients were hospitalized with cough, with three exhibiting productive sputum. Only one patient reported chills and myalgia. Computed tomography (CT) chest scans revealed unilateral pulmonary inflammation in all patients. Hematological analysis demonstrated markedly decreased retinol-binding protein and prealbumin levels across all four cases. Three patients exhibited elevated plateletcrit values. Additional abnormal laboratory parameters were observed in two patients, including neutrophilia, elevated high-sensitivity C-reactive protein, hyperfibrinogenemia, increased d-dimer, prolonged activated partial thromboplastin time, and elevated total bile acids, concurrent with lymphopenia and hypoalbuminemia. Serological testing for IgM antibodies against Legionella pneumophila and Mycoplasma pneumoniae yielded negative results in three patients. Throat swab specimens from two patients were analyzed for common respiratory pathogens, including influenza virus, parainfluenza virus, metapneumovirus, bocavirus, respiratory syncytial virus, coronavirus, rhinovirus, M. pneumoniae, and adenovirus. Patient 4 tested positive for adenovirus, while no other pathogens were detected in any patient. All patients recovered following treatment with azithromycin or doxycycline and were subsequently discharged.
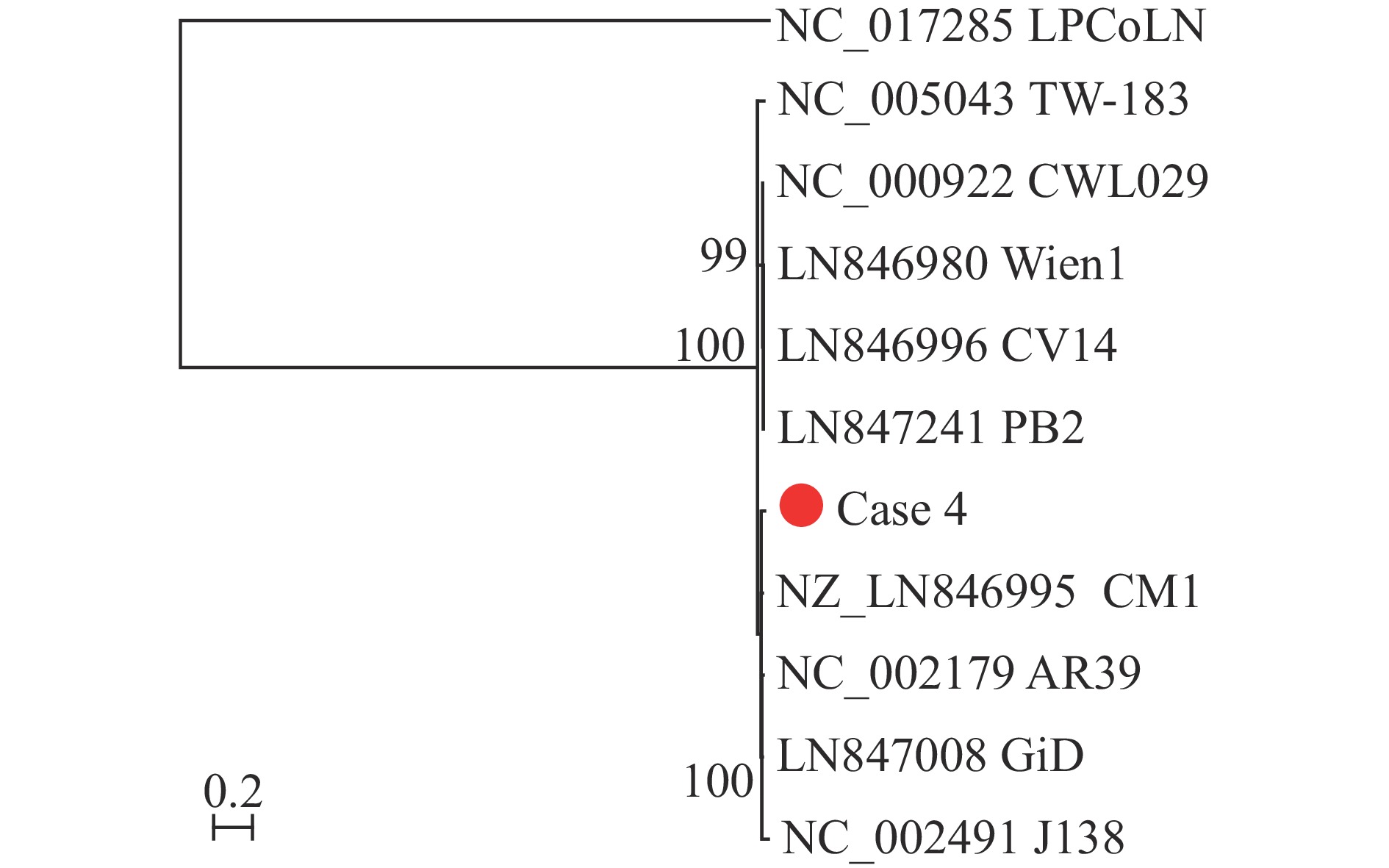
tNGS of bronchoalveolar lavage fluid samples from Patients 2 and 3 identified C. pneumoniae in Patient 2 (10,681 reads), while Patient 3 demonstrated a polymicrobial infection with C. pneumoniae (87 reads), Haemophilus influenzae (35,231 reads), and human adenovirus 5 (157 reads) (Table 1). Sanger sequencing confirmed C. pneumoniae infection, with 99%–100% homology to known C. pneumoniae 16S rRNA sequences and 98%–100% homology to the ompA gene. The sequences were deposited in GenBank (accession numbers PQ510372-PQ510375 for 16S rRNA, PQ522244-PQ522247 for ompA). Complete C. pneumoniae genomic sequences were attempted to be obtained using NGS-based hybrid capture methodology. Only Patient 4 yielded a sufficient genomic draft (GenBank BioProject PRJNA1177653), with phylogenetic analysis demonstrating close clustering with the prototype strain AR39 (Figure 1). MLST analysis revealed that all four patients were infected with sequence type 16 (ST16) (enoA:5, fumC:4, gatA:4, gidA:6, hemN:3, hflX:5, and oppA:7).
Characteristics Case #1 Case #2 Case #3 Case #4 Age (years)/sex 28/Female 36/Female 16/Male 14/Female Signs/symptoms 37.1 ℃
Productive cough37.5 ℃
Chills, Myalgia
Productive cough36.5 ℃
Productive cough38.5 ℃
Fever, Cough
Moist ralesIgM detection* (−) (−) / (−) Nucleic acid detection of common
respiratory pathogens†/ (−) / Adenovirus (+) tNGS / C. pneumonia
(10,681 reads)C. pneumonia
(87 reads)
Haemophilus influenzae
(35,231 reads)
Human Adenovirus 5
(157 reads)/ Blood test ↑ (APTT, D-dimer,
FIB, PCT, TBA)
↓ (PA, RBP)↑ (D-dimer, FIB, hs-CRP,
NEUT, PCT)
↓ (ALB, LYM,
PA, RBP)↑ (PCT, TBA)
↓ (ALB, PA, RBP)↑ (ADA, APTT, AST,
β-2-MG, CK, HBDH,
hs-CRP, LDH, NEUT)
↓ (LYM, PA, RBP)Note: “/” means the test was not performed; (−) means a negative result; ↑ and ↓ means increased and decreased test values, respectively.
Abbreviation: C. pneumoniae=Chlamydia pneumoniae; tNGS=Targeted next-generation sequencing; ADA=Adenosine Deaminase; ALB=Albumin; APTT=Activated partial thromboplastin time; AST=Aspartate aminotransferase; β-2-MG=Beta-2-microglobulin; CK=Creatine Kinase; FIB=Fibrinogen; HBDH=Alpha hydroxybutyrate dehydrogenase; hs-CRP=Hypersensitive C-reactive protein; LDH=Lactate dehydrogenase; LYM=Lymphocyte; NEUT=Neutrophil; PA=Prealbumin; PCT=Plateletcrit; RBP=Retinol-binding protein; TBA=Total bile acid.
* Simultaneous testing for IgM antibodies against Legionella pneumophila and Mycoplasma pneumoniae.
† Common respiratory pathogens include influenza virus, parainfluenza virus, metapneumovirus, bocavirus, respiratory syncytial virus, coronavirus, rhinovirus, Mycoplasma pneumoniae, and adenovirus.Table 1. Demographic and basic clinical features of the four patients.
-
In this study, we identified C. pneumoniae infection in four patients with severe pneumonia from Lishui City, Zhejiang Province, China. Analysis of the 16S rRNA and ompA gene sequences demonstrated 98% to 100% homology with known strains of C. pneumoniae. To further characterize the molecular epidemiology of these isolates, we sequenced seven housekeeping genes for MLST. Results revealed that all four patients were infected with the ST16 genotype. Based on currently available sequencing data for C. pneumoniae, MLST classification encompasses four distinct sequence types: ST16, ST17, ST18, and ST215. Among these, ST215 is exclusively derived from Australian koalas and Perameles bougainville, while the remaining three sequence types (ST16, ST17, and ST18) are associated with human infections (4–5).
All four patients with C. pneumoniae pneumonia initially presented with cough and fever of unknown origin, with productive cough observed in three cases. CT scans revealed unilateral pneumonia in all patients. Laboratory analyses demonstrated consistent abnormalities across all cases, notably decreased levels of retinol-binding protein and prealbumin. This finding represents a novel clinical observation not previously documented in the literature on C. pneumoniae infection (6–8). Both retinol-binding protein and prealbumin are hepatically synthesized proteins involved in vitamin A transport, and alterations in their concentrations serve as indirect indicators of systemic inflammatory responses (9–10). Therefore, monitoring these biomarkers in patients with C. pneumoniae pneumonia may provide valuable clinical insights, enabling physicians to better assess disease severity and potentially offering actionable biomarkers for clinical management.
C. pneumoniae frequently occurs as a co-infecting pathogen in pneumonia cases. In our cohort, two patients exhibited co-infections: one with a dual pathogen profile (adenovirus) and another with a triple pathogen profile (adenovirus and H. influenzae). Such co-infections typically present greater therapeutic challenges, underscoring the critical importance of accurate pathogen identification for targeted intervention. In this context, tNGS represents an increasingly valuable diagnostic modality for severe clinical infections, particularly for identifying complex co-infections. However, standard hospital pathogen detection protocols rarely include C. pneumoniae, highlighting how tNGS implementation could substantially enhance detection rates for this pathogen.
In conclusion, the epidemiology and predominant sequence types of C. pneumoniae in China remain inadequately characterized. While our findings contribute meaningful data to the existing literature, the limited sample size necessitates further comprehensive investigations into C. pneumoniae and other chlamydial infections.
-
The study was approved by the Ethics Committee of the National Institute of Communicable Disease Prevention and Control, China CDC (Approval no. ICDC-202115).
| Citation: |



 Download:
Download:




