-
Entering the first winter after the World Health Organization (WHO) declared coronavirus disease 2019 (COVID-19) no longer a Public Health Emergency of International Concern (PHEIC), an increase in acute respiratory infections caused by multiple respiratory pathogens resulted in increased hospitalizations. This rise in respiratory infections, particularly delayed pediatric Mycoplasma pneumoniae (M. pneumoniae), among specific age groups differed from typical seasonal patterns (1). These changes may be due to the immunity gap developed during the COVID-19 pandemic (2). This study investigated the current epidemiological trends of respiratory pathogens in Beijing, with a specific focus on pathogen composition. The findings, combined with global health recommendations, can offer valuable insights for addressing the ongoing challenges posed by respiratory infections in the future.
-
Oropharyngeal swabs, nasopharyngeal swabs, and sputum samples were collected from patients presenting with respiratory symptoms, including fever, cough, sore throat, shortness of breath, and other influenza-like symptoms. Samples were collected at Ditan Hospital, Haidian Hospital, and surrounding communities in Beijing from November 15, 2023, to early April 2024. The inclusion criteria consisted of individuals exhibiting at least one of the specified influenza-like symptoms and seeking medical care at the designated locations.
-
Total DNA/RNA was extracted from 200 μL of sample transport medium containing each patient biospecimen using the MagaBio Pathogens DNA/RNA Purification Kit (Catalog No. BSC75; BIOER) according to the manufacturer's instructions.
In total, 27 common respiratory pathogens were detected here, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), influenza viruses (IAV-IDV), human bocavirus (HBoV), human parainfluenza virus (HPIV-I-IV), respiratory syncytial virus (RSV), adenovirus (ADV), human metapneumovirus (HMPV), human rhinovirus (HRV), human coronaviruses (HCoV-NL63, HCoV-229E, HCoV-OC43, and HCoV-HKU1), and the bacterial pathogens Klebsiella pneumoniae (K. pneumoniae), Streptococcus pneumoniae (S. pneumoniae), Staphylococcus aureus (S. aureus), Legionella pneumophila (L. pneumophila), H. influenzae, Pseudomonas aeruginosa (P. aeruginosa), Acinetobacter baumannii (A. baumannii), M. pneumoniae, and M. catarrhalis, via multiplex quantitative PCR (qPCR) using specific primers, probes, and the detection kit (Influenza A virus, Influenza B virus, and SARS-CoV-2 Triple-Detection Kit, Catalog No. SKY-82130) (3). Lab-confirmed cases were defined as those with positive qPCR detection for the different pathogens.
-
Statistical analyses were performed using R (version 4.3.2). Descriptive statistics, including means, medians, and percentages, summarized the study population characteristics. Chi-square tests compared categorical variables, such as gender, pathogen detection rates between males and females, and differences in pathogen detection rates between hospital and community samples. Correlation analysis assessed interactions between different pathogens. The restricted cubic spline (RCS) method modeled the nonlinear relationship between age and the risk of pathogen occurrence. Multivariable logistic regression models, adjusting for confounders such as age, gender, and clinical symptoms, identified factors associated with pathogen positivity. All statistical analyses were conducted to ensure the reliability and validity of the results, and findings were reported alongside 95% confidence intervals (CIs). A P<0.05 was considered statistically significant.
-
Between November 2023 and April 2024, a total of 1,513 samples were collected from patients with respiratory tract infections from 2 sentinel hospitals (1,437) and surrounding communities (76) in Beijing. Of these samples, 770 were from male patients, and 743 were from female patients, resulting in a male-to-female ratio of 1.04:1.00. The positive rate for both males and females (χ2=0.84, P=0.40), as well as for hospital and community settings (χ2=3.53, P=0.06), did not show a significant difference. Additionally, there were 158 cases in children (<18 years old) and 1,355 cases in adults (≥18 years old), with a median age of 32 years. Among the collected samples, 787 (52.02%) tested positive for viruses. This included 429 cases (28.35%) of influenza virus infections, with 237 cases (55.24%) of IBV, 190 cases (44.29%) of IAV, and 2 cases (0.47%) of ICV. There were also 150 cases (9.91%) of SARS-CoV-2, 102 cases (6.74%) of HBoV, 72 cases (4.76%) of RSV, 42 cases (2.78%) of ADV, 20 cases (1.32%) of HMPV, and 13 cases (0.86%) of HRV infections. Regarding bacterial infections in patients with fever, 565 cases (37.34%) tested positive for bacteria. The highest detection rate was for H. influenzae, accounting for 15.07% of cases. The positive rates for other bacteria are shown in Table 1.
Pathogen type Pathogen name Wave 2 (%) Wave 3 (%) Sum (%) Viruses SARS-CoV-2 42 (5.41) 35 (6.36) 77 (5.81) Influenza A virus 170 (21.91) 19 (3.45) 189 (14.25) Influenza B virus 27 (3.48) 191 (34.73) 218 (16.44) Influenza C virus 1 (0.13) 1 (0.18) 2 (0.15) Influenza D virus 0 0 0 Human bocavirus 49 (6.31) 47 (8.55) 96 (7.24) Human metapneumovirus 5 (0.64) 11 (2.00) 16 (1.21) Respiratory syncytial virus 36 (4.64) 29 (5.27) 65 (4.90) Adenovirus 22 (2.84) 15 (2.73) 37 (2.79) Human rhinovirus 10 (1.29) 1 (0.18) 11 (0.83) Human parainfluenza virus I 22 (2.84) 48 (8.73) 70 (5.28) Human parainfluenza virus II 0 0 0 Human parainfluenza virus III 4 (0.52) 2 (0.36) 6 (0.45) Human parainfluenza virus IV 7 (0.90) 0 7 (0.53) Human coronavirus NL63 0 0 0 Human coronavirus 229E 10 (1.29) 0 10 (0.75) Human coronavirus OC43 2 (0.26) 0 2 (0.15) Human coronavirus HKU1 1 (0.13) 0 1 (0.08) Bacterial Klebsiella pneumoniae 67 (8.63) 32 (5.82) 99 (7.45) Streptococcus pneumoniae 65 (8.38) 31 (5.64) 96 (7.24) Staphylococcus aureus 50 (6.44) 13 (2.36) 63 (4.75) Legionella pneumophilia 1 (0.13) 0 1 (0.08) Haemophilus influenzae 123 (15.85) 71 (12.91) 194 (14.63) Pseudomonas aeruginosa 50 (6.44) 27 (4.91) 77 (5.81) Acinetobacter baumannii 53 (6.83) 16 (2.91) 69 (5.20) Moraxella catarrhalis 19 (2.45) 6 (1.09) 25 (1.89) Mycoplasma pneumoniae 25 (3.22) 8 (1.45) 33 (2.49) Note: Wave 2 spans from October 10 to December 25, 2023, while Wave 3 extends from December 25, 2023 to February 10, 2024.
Abbreviation: SARS-CoV-2=severe acute respiratory syndrome coronavirus 2; qPCR=quantitative polymerase chain reaction.Table 1. The number of positive cases and the percentage of 27 respiratory pathogens identified by qPCR in outpatients.
Based on prevalence trends of dominant pathogens, this period can be divided into four epidemic waves: the pre-winter M. pneumoniae outbreak with declining SARS-CoV-2 prevalence (Wave 1), followed by the emergence of the IAV epidemic from October to the end of December 2023 (Wave 2), in which the 5.41% detection rate of SARS-CoV-2 was lower than during the COVID-19 pandemic. In contrast to the pathogen prevalence at the end of 2023, the detection rate of IBV increased rapidly and became the dominant pathogen in early 2024 (Wave 3), reaching 34.73%, and then gradually decreased in early February. By mid-February 2024, SARS-CoV-2 infections began to rise, reaching a peak detection rate of 39.04% in mid-March, then gradually decreasing in April, with the number of outpatients with respiratory diseases decreasing significantly (Wave 4, Figure 1). These four waves, each with a different dominant pathogen, highlight the dynamic nature of pathogen prevalence and emphasize the importance of continuous monitoring.
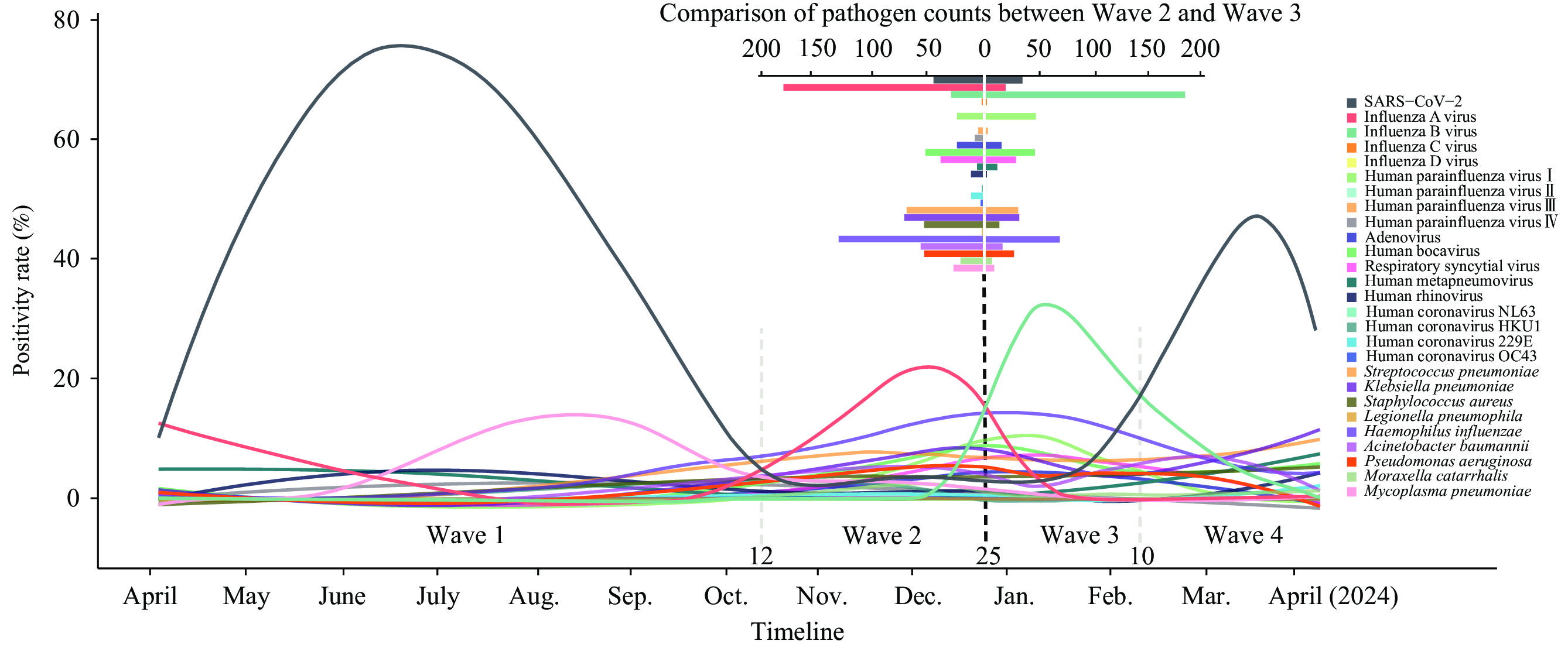 Figure 1.
Figure 1.Epidemic trend of each pathogen detected by qPCR (November 2023 to April 2024).
Note: This graph depicts the epidemic trend of various pathogens detected by qPCR from November 2023 to April 2024. Each colorful trajectory represents a different pathogen, with the trends reflecting changes in their detected levels over time. The graph's mirrored layout, with an axis on 25 December, facilitates a comparison of trends in Wave 2 and Wave 3. In the upper right, the bar graph details the distribution of detection count for each pathogen during the study period.
Abbreviation: SARS-CoV-2=severe acute respiratory syndrome coronavirus 2; qPCR=quantitative polymerase chain reaction.
-
Of the 1,513 cases of respiratory pathogen infection, 284 (18.77%) were co-infected with more than two respiratory pathogens. Among these, H. influenzae was frequently co-infected with other viral and bacterial pathogens (66 cases, 4.36%), followed by IBV (64 cases, 4.23%) and IAV (40 cases, 2.64%). Notably, 18 cases (1.19%) were co-infected by two viruses, including 13 cases of IAV/IBV and HBoV and 5 cases of HBoV and HPIV-I (Figure 2). Correlation analysis revealed potential interactions and impact patterns among these pathogens (Figure 3). A significant negative correlation was found between SARS-CoV-2 and influenza viruses, particularly IBV (P<0.0001). H. influenzae was more likely to co-infect with other pediatric-prevalent pathogens, such as ADV (P<0.05), HMPV (P<0.05), and HRV (P<0.0001), but exhibited a significant negative correlation with SARS-CoV-2 (P<0.0001). Additionally, M. catarrhalis showed positive correlations with coronaviruses, including SARS-CoV-2 (P<0.0001) and HCoV-229E (P<0.0001). Furthermore, HBoV and HPIV-I showed a significant positive correlation. Interestingly, influenza A and B viruses had a relatively low likelihood of co-infection with other respiratory viruses.
 Figure 2.
Figure 2.Overview of single and co-infections with different pathogens in Beijing (November 2023 to April 2024).
Note: The figure illustrates the co-infection patterns of various pathogens. The horizontal bars represent the total number of infections for each pathogen. The vertical bars display the distribution of infections observed, including both single infections (each circle) and co-infections (linked circles). Only co-infections involving at least five cases are shown.
Abbreviation: SARS-CoV-2=severe acute respiratory syndrome coronavirus 2.
 Figure 3.
Figure 3.Co-infection and correlation among different pathogens in Beijing (November 2023-April 2024).
Note: The heatmap depicts the co-infection and correlation patterns among various pathogens. The color intensity in each cell represents the correlation coefficient between pairs of pathogens: red for positive correlation and blue for negative correlation, with darker shades indicating stronger relationships. The presence of asterisks within the cells denotes statistical significance.
Abbreviation: SARS-CoV-2=severe acute respiratory syndrome coronavirus 2.
* P<0.05;
** P<0.01;
*** P<0.001;
**** P<0.0001.
-
The nonlinear relationship between respiratory pathogens and outpatient age was modeled and visualized using RCS. This analysis revealed a significant nonlinear relationship between age and pathogen occurrence risk, with particularly pronounced changes in hazard ratios (HR) observed at certain age intervals (Figure 4). Generally, the infection risk profiles of SARS-CoV-2 (Figure 4A), IAV (Figure 4B), and IBV (Figure 4C) exhibited a similar pattern. Susceptibility gradually increased in patients under 30 years old, peaked in the 30–40-year age group, and then decreased. However, unlike SARS-CoV-2 and IAV, which mainly infected middle-aged and elderly individuals, IBV primarily infected middle-aged individuals, with the infection risk rapidly declining after 40 years old. In contrast, H. influenzae (Figure 4H) showed increased susceptibility among adolescents, with a substantial risk decrease as age advanced, ultimately stabilizing. HBoV (Figure 4E), RSV (Figure 4F), A. baumannii (Figure 4I), P. aeruginosa (Figure 4J), K. pneumoniae (Figure 4K), and S. aureus (Figure 4L) exhibited a U-shaped relationship with age. Among individuals under 30 years old, the risk of infection with these pathogens declined with advancing age, reaching the lowest risk among teenagers and young adults but increasing thereafter with further aging, demonstrating an elevated susceptibility trend among the elderly population. For S. pneumoniae (Figure 4G) and other pathogens, the lack of significant nonlinear age-related patterns could be attributable to insufficient statistical power due to sample size limitations or an inherent lack of strong age-dependent susceptibility patterns for these pathogens in the study population.
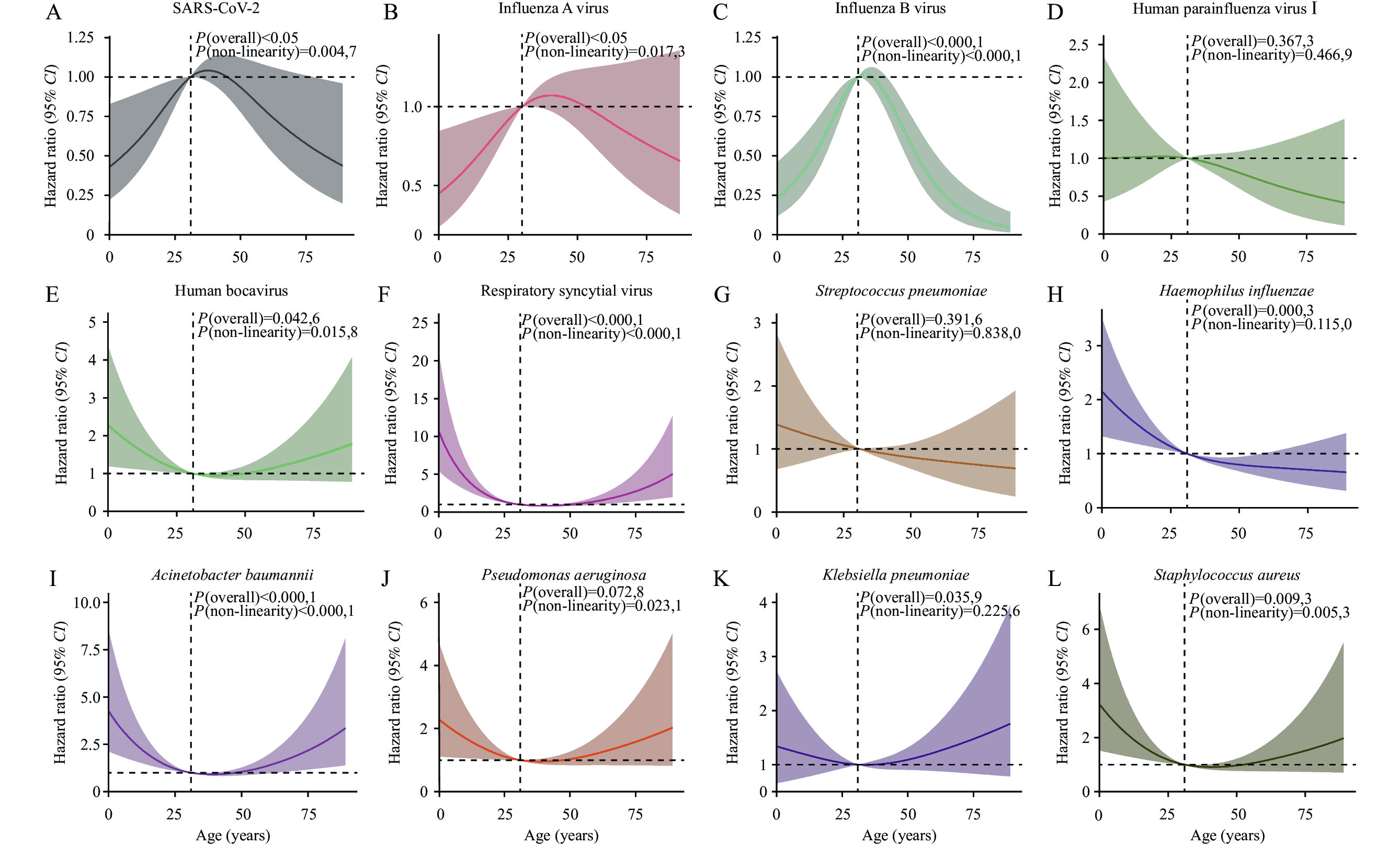 Figure 4.
Figure 4.Association of respiratory pathogens and the age of outpatients.
Note: The graphs present the nonlinear association analysis between pathogens (counts>50) and age, modeled using the RCS method. Solid lines represent the HRs of the influence of age on the occurrence of pathogens, while the shaded areas indicate 95% CIs. Knot locations are automatically selected based on the quantiles of age distribution to reveal the nonlinear trend of pathogen risk with age. Additionally, the P-values for overall association and nonlinearity are provided for interpretation.
Abbreviation: SARS-CoV-2=severe acute respiratory syndrome coronavirus 2; RCS=restricted cubic spline; HR=hazard ratio; CI=confidence interval.
-
This study focused on pathogen patterns in outpatients with respiratory infections from November 2023 to April 2024. The resurgence of various non-SARS-CoV-2 respiratory pathogens has fluctuated considerably, but the pathogen species remained unchanged. Several epidemic waves occurred, beginning with M. pneumoniae, followed by IAV and then IBV. By the end of February 2024, a new wave of SARS-CoV-2 re-emerged and peaked in mid-March. These findings indicate a resurgence of epidemic dynamics for respiratory pathogens from November 2023 to April 2024, with SARS-CoV-2 potentially persisting in long-term circulation in humans, similar to influenza virus and other seasonal pathogens. Notably, SARS-CoV-2 and influenza viruses appeared in staggered epidemic waves, suggesting that immune-mediated interference might cause one virus to diminish during the peak of another, thereby influencing their respective prevalence patterns (4). However, the exact epidemic patterns of these pathogens require further monitoring.
Previous studies have shown that multiple respiratory pathogens can easily infect different age groups (1). SARS-CoV-2 commonly infects adults, while its detection rate in children is low (5), and pediatric infections present with mild, short-lived symptoms (6). The infection risk profiles of IAV and IBV showed a similar pattern to SARS-CoV-2, with susceptibility peaking in the 30–40-year age group. However, a notable increase in pediatric-prevalent pathogens has been observed this winter. It began with a significant outbreak of M. pneumoniae among children. As the season progressed, alternating waves of IAV and IBV were observed, leading to a gradual decrease in the risk of infections with HBoV, RSV, A. baumannii, K. pneumoniae, S. aureus, and P. aeruginosa in adults. This risk reduction was most pronounced among adolescents and middle-aged adults. However, the risk subsequently increased with advancing age, particularly in the elderly, indicating increased susceptibility in this population.
The data from this study showed that the co-infection of viruses and/or bacteria increased the complexity of respiratory pathogen infections during this epidemic season compared to the COVID-19 pandemic period. The likelihood of co-infection with other respiratory viruses is relatively low for influenza A and B viruses, which may reflect specific host-pathogen interactions or immune responses (7). However, there was an elevated risk of secondary bacterial infections following viral respiratory illnesses. Co-infections of viral and bacterial pathogens were important factors that prolonged the infection process, highlighting the need for comprehensive diagnostic approaches to guide effective treatment strategies. In addition to IAV and IBV, other respiratory pathogens, such as RSV, ADV, HRV, and various bacteria, were frequently detected during the epidemic waves. Studies indicate that during influenza epidemics, co-infections involving multiple bacterial species are also common, with nearly 55.6% of severe influenza patients in ICUs experiencing exacerbated conditions due to co-infections with bacteria such as S. pneumoniae, S. aureus, M. pneumoniae, and H. influenzae (8–9). The rise of antibiotic resistance inevitably makes these co-infections a significant cause of severe pneumonia. In addition, the drug resistance-related genes can also be exchanged among bacteria, which further complicates the treatment. Therefore, co-infection among viruses, as well as between viruses and bacteria, is a major concern (10), especially bacteria that show resistance to commonly used antibiotics.
In this study, 27 causative agents for respiratory infections were detected. The results can enhance the understanding of epidemic dynamics and inform effective control strategies. Despite some limitations, such as samples being collected from only 2 sentinel hospitals in the northwest and northeast of Beijing rather than citywide and the relatively small number of pediatric cases, which may introduce bias. Nonetheless, these findings highlight the complexity of respiratory pathogen infections and the importance of comprehensive surveillance.
The alternating epidemics of influenza and SARS-CoV-2 increase the difficulty of prevention and control. The "immunity gap" caused by reduced exposure to common pathogens during prolonged lockdowns may lead to a temporary decline in population immunity, increasing individuals' susceptibility to infections (2). This phenomenon underscores the need for strategic vaccination campaigns (11), particularly for pathogens that frequently co-infect with other agents, such as influenza (12). To reduce the risk of widespread outbreaks, developing more convenient, accurate, and efficient detection methods is crucial. These advancements will significantly improve disease surveillance and control measures in the future.
HTML
Specimen Source
Pathogens Detection
Statistical Analysis
Epidemic Characteristics of the Respiratory Pathogens
Co-infection Patterns of the Respiratory Pathogens
Age Distribution of the Respiratory Pathogens
| Citation: |



 Download:
Download:




