-
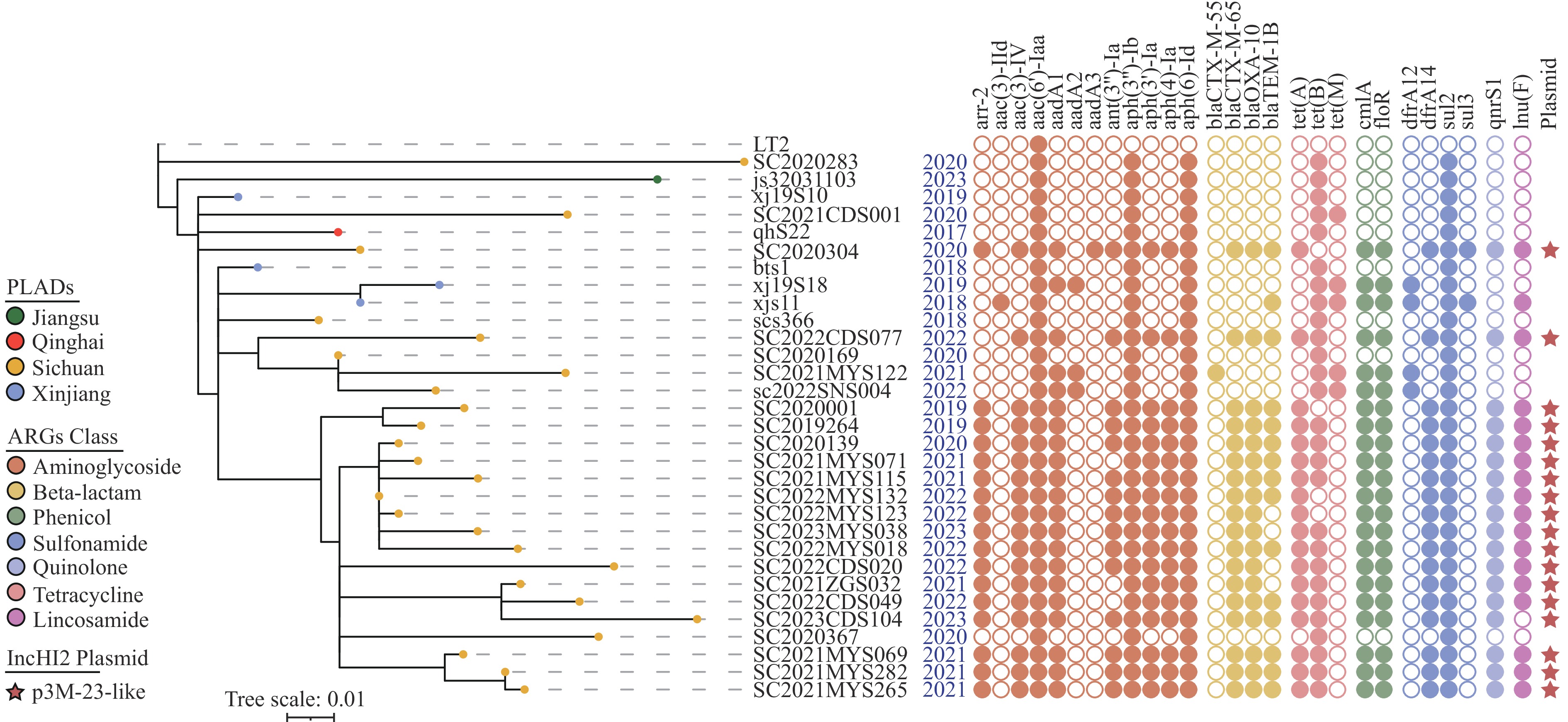
A total of 31 Salmonella 4,[5],12:i:- sequence type (ST) 8333 isolates, primarily obtained from sporadic diarrhea cases between 2017 and 2023, were identified in the National Molecular Tracing Network for Foodborne Disease Surveillance (TraNet) database based on early-warning information from the TraNet module. These isolates were mainly sourced from Sichuan Province (n=25). Phylogenetic analysis based on whole-genome single nucleotide polymorphisms (wgSNPs) revealed a close genetic relationship (1–27 SNPs) among these isolates, indicating that the ST8333 strain spread to Xinjiang, Sichuan, and Jiangsu provincial-level administrative divisions (PLADs) after originating in Qinghai Province in 2017. A total of 27 antimicrobial resistance genes (ARGs) belonging to seven classes were identified, with strains carrying blaCTX-M-55, blaCTX-M-65 or blaOXA-10 all isolated from Sichuan Province. Nanopore sequencing characterized an IncHI2 plasmid like p3M-23, which was recovered from swine fecal swabs on a swine farm in Sichuan Province. These plasmids carried multiple resistance genes, suggesting that ST8333 acquired multidrug-resistance plasmids horizontally during its spread in Sichuan Province, which in turn further contributed to the regional spread of these isolates. The continued detection of ST8333 isolates indicates their ongoing spread. Our findings suggest the imperative need to implement monitoring and control measures to prevent further spread of the multidrug-resistant ST8333 to other PLADs.
In China, over 900 sentinel hospitals across 31 PLADs and Xinjiang Production and Construction Corps (XPCC) conduct active surveillance annually. Approximately half of foodborne outbreaks are caused by bacterial pathogens, with Salmonella being the second most common pathogen in these outbreaks (1). Since 2009, S. 4,[5],12:i:- has become one of the four most predominant serotypes causing human salmonellosis in China. Previous studies have reported that ST34 S. 4,[5],12:i:- is the most prevalent in the country (2–3). Surveillance data from TraNet indicate that ST34 is the primary sequence type among S. 4,[5],12:i:- isolates from diarrhea cases in China, followed by ST19.
From 2017 to 2023, active surveillance yielded over 1,000,000 fecal specimens with an average Salmonella detection rate of 5.77%. In late 2023, the TraNet early-warning module, using whole-genome sequencing (WGS) data, identified a new regionally prevalent sequence type, ST8333. Active monitoring indicated 31 ST8333 isolates from sporadic diarrhea cases (2017–2023) were submitted to the TraNet Salmonella database. These isolates originated primarily from four PLADs: Sichuan (n=25), Xinjiang (n=4), Qinghai (n=1), and Jiangsu (n=1). The average prevalence of ST8333 among S. 4,[5],12:i:- isolates from diarrhea patients in Sichuan was 3.32%. Serotype and antimicrobial resistance gene (ARG) analyses were performed using the TraNetGenotype plugin. wgSNP analysis was performed using TraNetCE. Plasmid contigs were predicted using Platon (version 1.7) (4).
Based on the isolation date, the S. 4,[5],12:i:- sequence type (ST) 8333 was first identified in a patient with diarrhea in Qinghai Province and subsequently detected in Xinjiang, Sichuan Province, and Jiangsu Province. A total of 27 antimicrobial resistance genes (ARGs) belonging to seven classes were identified, with no mutations detected in the quinolone resistance-determining region. The prevalence of ARGs conferring resistance to aminoglycosides, tetracyclines, and sulfonamides was 100%. The prevalence of ARGs, including blaCTX-M-55, blaCTX-M-65, blaOXA-10, and blaTEM-1B, from beta-lactam was 61.29% (19/31). Strains carrying blaCTX-M-55, blaCTX-M-65 or blaOXA-10 were all isolated from Sichuan Province. Plasmid prediction indicated that these three ARGs were located on plasmid-like contigs.
To identify the possible plasmid, we used the predicted plasmid contigs as a query sequence in the NCBI BLASTn tool. An IncHI2 plasmid, named p3M-23 (OP970992), in S. 4,[5],12:i:- strain recovered from swine fecal swabs in Sichuan Province, was found to be similar to the query plasmid contigs. A total of 18 strains, all sourced from Sichuan Province, could be mapped to p3M-23 using the short sequencing reads. SC2019264 was selected for nanopore sequencing to obtain the plasmid sequence. Assembly results showed that SC2019264 carried an IncHI2 plasmid named pSC2019264 (Figure 1). The size of pSC2019264 was 246,828 bp. Compared with p3M-23, pSC2019264 acquired blaTEM-1 region.
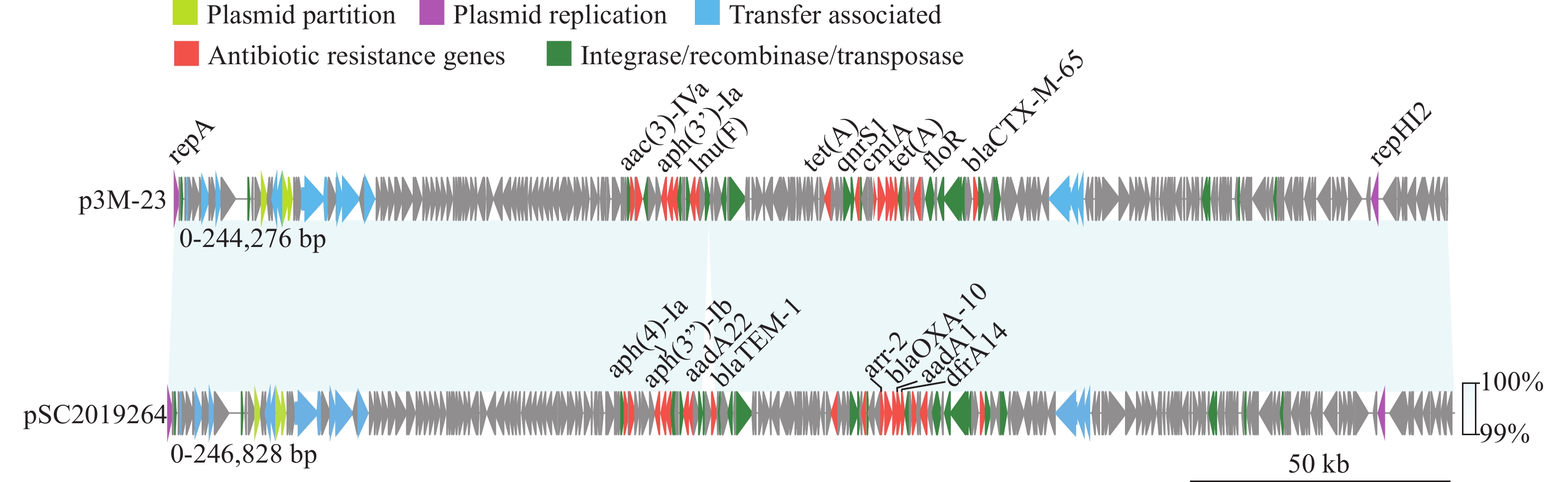 Figure 1.
Figure 1.Linear plasmid sequence comparison between p3M-23 and pSC2019264.
Note: Pistachio denotes the IncHI plasmid conjugative transfer partition protein, orchid represents the IncHI-type plasmid replication initiator protein, and sky blue signifies the IncHI-type conjugal transfer protein. Antimicrobial resistance genes and transposase or integrase genes are in red and green, respectively.To study the evolutionary relationships of ST8333 isolates, we constructed a wgSNP phylogenetic tree using S. Typhimurium LT2 (GenBank accession No.: NC_003197) as the reference sequence. The wgSNP analysis revealed that the number of SNP differences among the 31 strains ranged from 1 to 27, indicating that these ST8333 strains recently evolved from a common ancestor. The evolutionary analysis indicates that since the isolation of qhS22 in 2017, ST8333 isolates have spread to Xinjiang, Sichuan, and Jiangsu PLADs. Combined with the plasmid sequence analysis results, we infer that ST8333 acquired a multidrug-resistant plasmid like p3M-23 through horizontal transfer and continued to spread locally after reaching Sichuan in 2018 (Figure 2).
-
Since S. 4,[5],12:i:- was first discovered in the late 1980s, it has attracted widespread attention globally. Previous studies have found a strong association between S. 4,[5],12:i:-, pig breeding, and the consumption of contaminated pork (5–7). In Europe, pigs are the primary source of foodborne S. 4,[5],12:i:-, making it one of the three most common serotypes in human cases (8). In the US, S. 4,[5],12:i:- is the most prevalent serotype in market swine and the third most common in pork products (9).
Pork consumption may be a source of ST8333 infection in Sichuan Province. In this study, 80.65% (25/31) of ST8333 isolates were from Sichuan Province. Sichuan, as one of the major PLADs in southern China, has the highest reported cases of human diarrhea caused by S. 4,[5],12:i:-. As the PLAD with the largest volume of pigs for slaughter, we speculate that Sichuan Province provides conditions for the survival and spread of ST8333 or new types of S. 4,[5],12:i:- that may emerge in the future.
Plasmid analysis revealed high similarities between plasmid pSC2019264 and the known multidrug-resistance plasmid p3M-23. The presence of these plasmids in ST8333 isolates suggests potential horizontal gene transfer events contributing to the acquisition of multidrug resistance. More than half of the ST8333 isolates carry a p3M-23-like plasmid, originally sourced from a swine fecal swab. This further suggests that the ST8333 strain isolated from the patients in this study may be related to the consumption of contaminated pork products. Additionally, 61.29% of ST8333 isolates carry the extended-spectrum β-lactamase (ESBL) gene blaCTX-M-55 or blaCTX-M-65, which commonly mediate resistance to third-generation cephalosporins. Their presence poses a significant challenge in clinical settings, as it limits treatment options and contributes to the spread of antimicrobial resistance, leading to treatment failure. The prevalence of ESBL-producing strains is nearly five times higher among ST8333 isolates in this study compared to the 13.00% in ST34 strains previously reported (10).
Our wgSNP analysis demonstrated a genetic relationship between the original 2017 Salmonella ST8333 isolate from Qinghai and isolates from other PLADs, indicating continued circulation of ST8333 isolates within the community. We also observed a distinct cluster with 3–19 SNP differences among isolates carrying the p3M-23-like plasmid. The inference of horizontal transfer events, supported by plasmid sequence analysis, underscores the need for effective measures to prevent the dissemination of multidrug-resistant ST8333 strains. To prevent a nationwide pandemic, food production enterprises and public health departments should prioritize surveillance of these ST8333 isolates. Our study highlighted Sichuan as a reservoir for the emergence and transmission of multidrug-resistant ST8333 strains, emphasizing the need for strengthened surveillance in this region to rapidly interrupt transmission.
This study was subject to some limitations. First, only isolates from sporadic diarrhea cases were investigated, isolates from food animals or food products were not included. Second, not all clinical isolates before 2017 were subjected to WGS analysis, earlier ST8333 isolates might exist.
In conclusion, this study underscores the urgent need for surveillance and control measures to mitigate the spread of multidrug-resistant ST8333 isolates, particularly in regions with high prevalence. Strategies for monitoring antimicrobial resistance and implementing targeted interventions are essential to safeguard public health and mitigate the impact of emerging bacterial pathogens like ST8333.
-
The invaluable contribution of all members in all participant CDCs and sentinel hospitals for their dedication to foodborne disease investigation, reporting and data auditing.
HTML
| Citation: |



 Download:
Download: