-
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant emerged in November 2021 (1). Within a month, Omicron subvariants were found in varying proportions in different regions worldwide and rapidly replaced the previous variants of concern (VOCs) (2-3). Studies have reported that the transmissibility and neutralization evasion capability of Omicron are enhanced in comparison to prior strains (4-5). The wide spread of the Omicron variant across the world has led to multiple recombinants emerging and significant intra-Omicron evolution. Since November 14, 2022, Omicron has evolved into 661 lineages, including recombinants (6). Some of them, designated as Omicron subvariants under monitoring by the World Health Organization (WHO) (7), have enhanced transmission advantages and have become more difficult to prevent and control than ever before.
In the mainland of China, several domestic transmissions associated with imported Omicron infections began to emerge in December 2021 (8-11), posing a significant challenge to the dynamic zero-coronavirus disease 2019 (COVID-19) policy. Under the current Omicron wave, genomic surveillance for SARS-CoV-2 from imported COVID-19 cases has had a crucial role in tracking the variant’s international spread and inferencing the origin of domestic transmissions. According to the Protocol on Prevention and Control of COVID-19 (Edition 8), the laboratories of provincial CDCs were required to conduct SARS-CoV-2 whole-genome sequencing for samples from all imported COVID-19 cases and submit the genomic sequences to the China CDC. This study analyzed genomic surveillance data from January to June 2022 to understand the importation of SARS-CoV-2 variants in the mainland of China during the Omicron epidemic.
-
The sequences of SARS-CoV-2 from imported cases were submitted from provincial CDC laboratories to the national China CDC for verification and further analyses. Sequences involved in the study were submitted from January to July 2022 — with a collection date before June 30, 2022. Data from Hong Kong Special Administrative Region (SAR), China; Macao SAR, China; and Taiwan, China were not included in this study. Global SARS-CoV-2 sequence surveillance data in the Global Initiative on Sharing All Influenza Data (GISAID) were extracted from the COV-spectrum platform on July 17, 2022 (12).
Consensus sequences were assessed for quality control by the Nextclade web tool (https://clades.nextstrain.org; version 2.4.1) with parameters for missing data, mixed sites, private mutations, mutation clusters, frameshifts, and stop codons. Sequences with good quality control status were typed using the Phylogenetic Assignment of Named Global Outbreak Lineages (PANGOLIN; version 4.0.5) web tool (13). The SARS-CoV-2 VOCs and Omicron subvariants under monitoring were classified according to the WHO’s designation (7).
-
A total of 4,946 SARS-CoV-2 genomic sequences collected before June 30, 2022, from imported COVID-19 cases were submitted to the China CDC during the period of January to July, 2022. Except the Xizang (Tibet) Autonomous Region, Xinjiang Uygur Autonomous Region, Qinghai Province, and Anhui Province, the rest of the provincial-level administrative divisions (PLADs) had sequences submitted in various amounts (Figure 1A). To validate sequence submissions' timeliness, each sequence's deposition time was calculated according to the date of sample collection and the date of sequence submission. The results showed that Shanghai Municipality had the longest median time of sequence deposition, with a 28-day median time (IQR=21–38), followed by Liaoning Province with a 26-day median (IQR=11–44). The median time of sequence deposition of the remaining PLADs was within 3 weeks. However, 15, 10, and 6 sequences were submitted 90 days after sample collection in Yunnan Province, Henan Province, and Shanghai, respectively (Figure 1B).
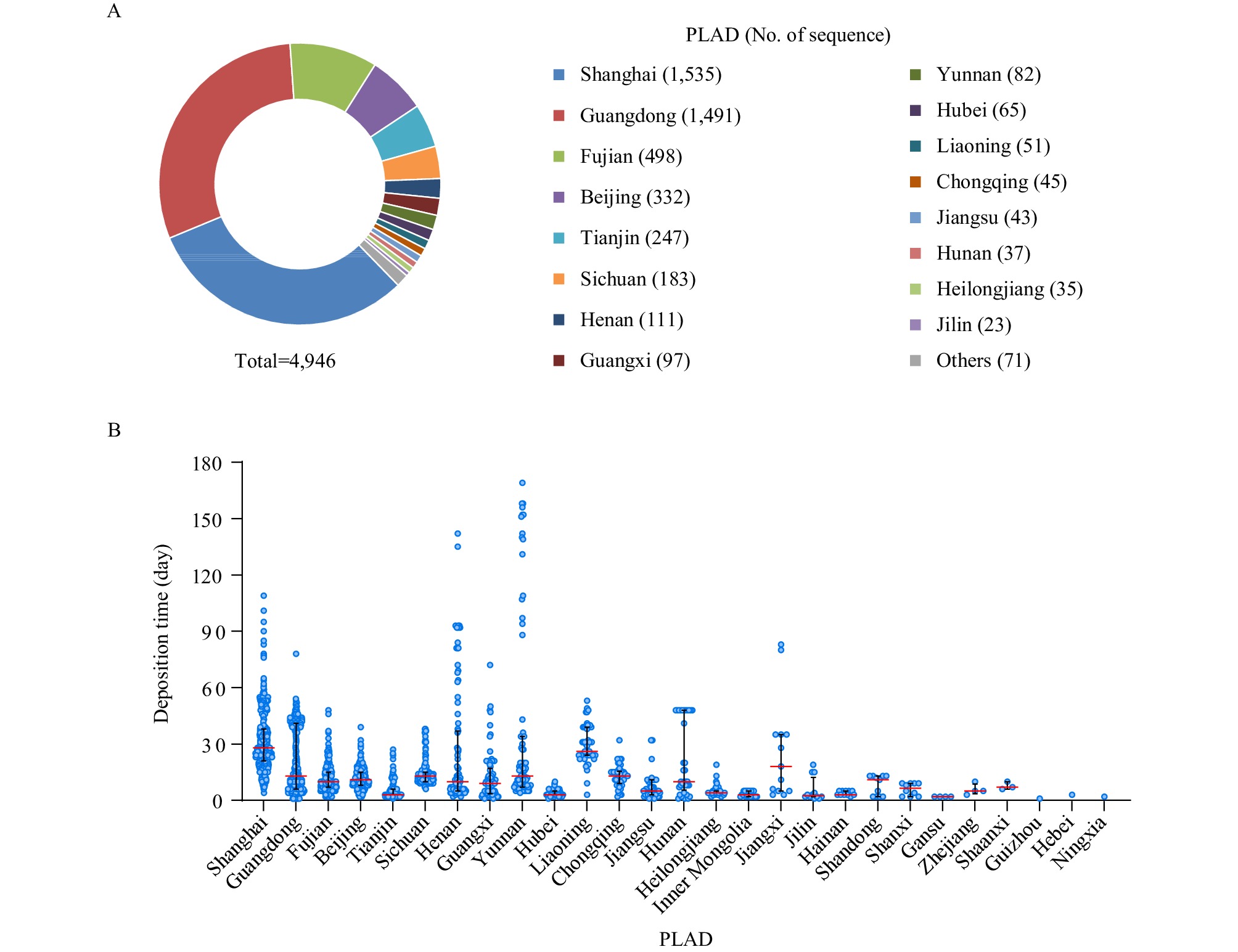 Figure 1.
Figure 1.The surveillance of SARS-CoV-2 genomes in imported cases in the mainland of China from January to June 2022. (A) The number of genomic sequences of imported cases submitted by PLAD. (B) The sequence deposition time of each PLAD from sample collection to sequence submission.
Note: In panel A, the 11 PLADs with less than 10 sequence submissions were merged in the category “Others”. In panel B, the PLADs were listed in order of the number of sequence submissions from most to least. Data was shown as median with interquartile range. Data as of July 31, 2022.
Abbreviation: SARS-CoV-2=severe acute respiratory syndrome coronavirus 2; PLAD=provincial-level administrative division.
Of the 4,946 sequences obtained, 4,742 were collected from January to June 2022. Among the 4,742 sequences, 3,893 (82.10%) had good quality control status, allowing for further lineage analysis (Figure 2A). Of them, the Omicron variant accounted for 98.92% (3,851/3,893), including 5 major lineages: BA.1, BA.2, BA.4, BA5, and recombinant. In January, the proportion of BA.1 was the highest: around 86.8%. Subsequently, BA.2 replaced BA.1, becoming dominant in February (79.9%) and overwhelmingly dominant from March to May (more than 90% per month). BA.4 and BA.5 were first detected in April, and their proportion gradually increased, accounting for 11.2% and 41.9% in June, respectively. In the lineages of Omicron, one recombinant XU, a hybrid of BA.1 and BA.2, and four subvariants under monitoring (BA.4, BA.5, BA.2.12.1, and BA.2.13) were recorded in the imported SARS-CoV-2 surveillance (Table 1). Three of them (BA.4, BA.5, and BA.2.12.1) had caused local transmissions in the mainland of China, and the time of causing local transmission was at least 26 days later than that recorded in the surveillance.
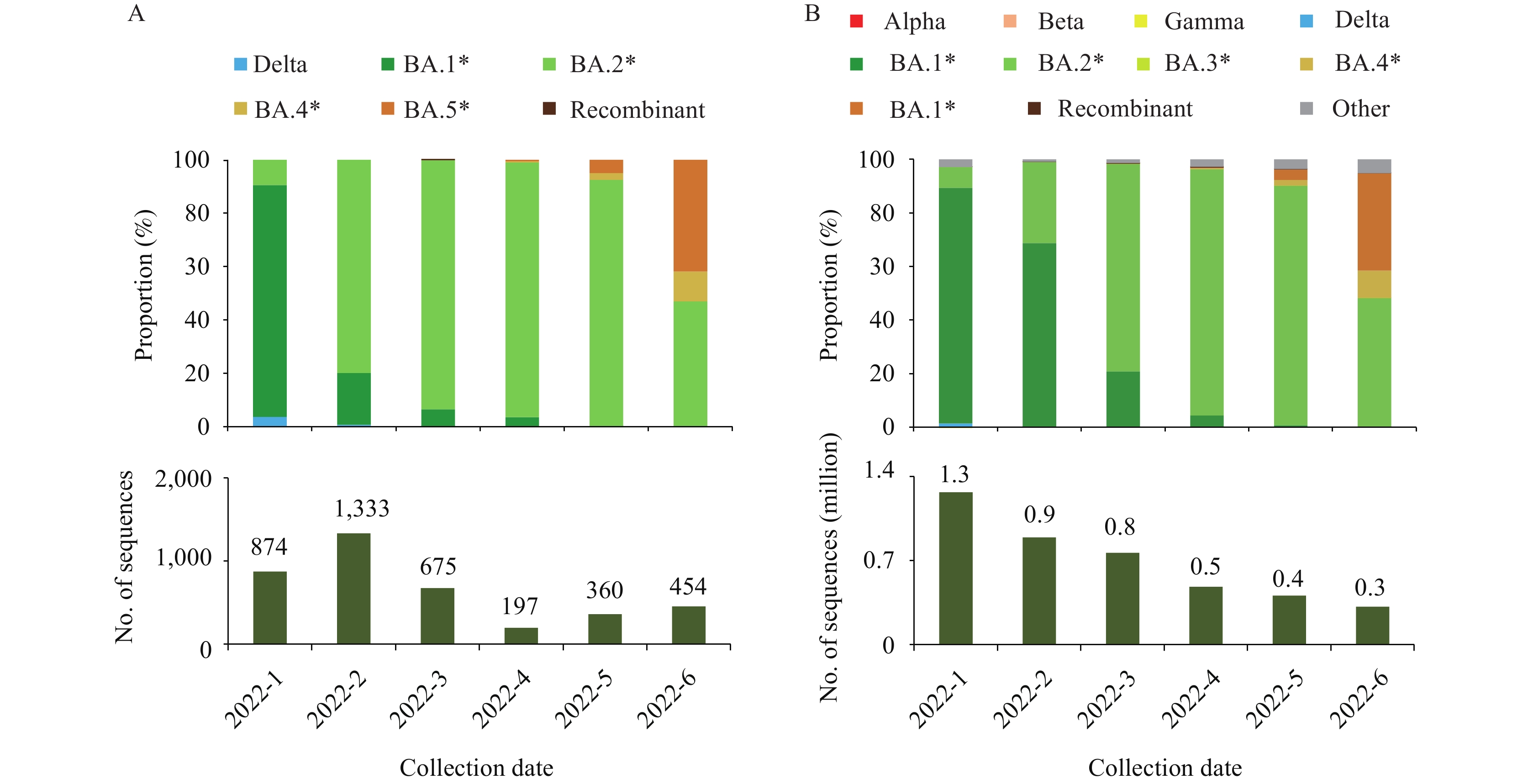 Figure 2.
Figure 2.The proportion of SARS-CoV-2 VOCs in the imported SARS-CoV-2 surveillance and in GISAID from January to June 2022. (A) The imported SARS-CoV-2 surveillance in the mainland of China (Data as of July 31, 2022). (B) GISAID (Data as of July 17, 2022).
Note: Recombinant includes BA.1/BA.2 circulating recombinant forms such as XE. The catalogs BA.1*, BA.2*, BA.3*, BA.4*, BA.5* and Recombinant all belong to Omicron.
Abbreviation: SARS-CoV-2=severe acute respiratory syndrome coronavirus 2; VOC=variant of concern; GISAID=the Global Initiative on Sharing All Influenza Data.
* Includes the descendent lineages.
Lineage* Earliest recorded sequence in the imported SARS-CoV-2 surveillance Date of first local transmission Earliest documented samples in the world† Submitted PLAD Submission date Imported country XU§ Guangdong 2022-4-21
(Collection date: 2022-3-12)United Arab Emirates − Japan,
2022-1BA.4¶ Guangdong 2022-5-4
(Collection date: 2022-4-30)Netherlands Guangdong,
2022-6-25South Africa,
2022-1BA.5¶ Shanghai 2022-5-15
(Collection date: 2022-4-29)Uganda Chongqing,
2022-6-11South Africa,
2022-1BA.2.12.1¶ Sichuan 2022-4-29
(Collection date: 2022-4-15)Canada Sichuan,
2022-7-15United States of America,
2021-12BA.2.13¶ Guangdong 2022-6-24
(Collection date: 2022-6-20)Canada − United States of America,
2021-12Abbreviation: SARS-CoV-2=severe acute respiratory syndrome coronavirus 2; PLAD=provincial-level administrative division; WHO=World Health Organization.
* Includes the descendent lineages.
† According to the data published by the Pango lineages (6).
§ Recombinant lineage of BA.1 and BA.2.
¶ Omicron subvariants under monitoring designated by WHO (7).Table 1. The detection of SARS-CoV-2 Omicron recombinant and subvariants under monitoring in the mainland of China and the world.
The distribution of dominant lineages for the sequences collected from imported cases in the mainland of China was in accordance with the worldwide sequencing data obtained from GISAID (Figure 2B). However, there were differences in the diversity and percentage of the lineages. The Alpha, Beta, Gamma, and Omicron BA.3 subvariants were recorded in the first half of 2022 in GISAID but were not detected in the imported SARS-CoV-2 surveillance in the mainland of China. The percentage of BA.2 in the imported SARS-CoV-2 surveillance was far more than that in GISAID (February: 79.9% vs. 30.2%; March: 93.3% vs. 77.4%). Notably, the number of sequences shared in GISAID decreased month by month in the first half of 2022.
To gain insight into the introduction of Omicron into the mainland of China, this study generated a Sankey diagram to illustrate the relationships between the origin and destination of each Omicron imported case (Figure 3A). The results showed that 3,851 Omicron cases came from 118 countries or regions. The four major lineages, BA.1, BA.2, BA.4, and BA.5, were imported from 89, 81, 23, and 40 countries or regions, respectively. Hong Kong imported the most significant number of cases (32.30%, 1,244/3,851) and mainly introduced them to Guangdong Province (49.84%, 620/1,244) and Shanghai (32.56%, 405/1,244). Furthermore, almost all the imported cases from Hong Kong from January to June 2022 were the BA.2 strain (98.71%, 1,228/1,244). Finally, the sequences of the imported cases reported from Shanghai and Guangdong had the widest breadth of origin locations: sourcing from 73 and 65 countries or regions, respectively.
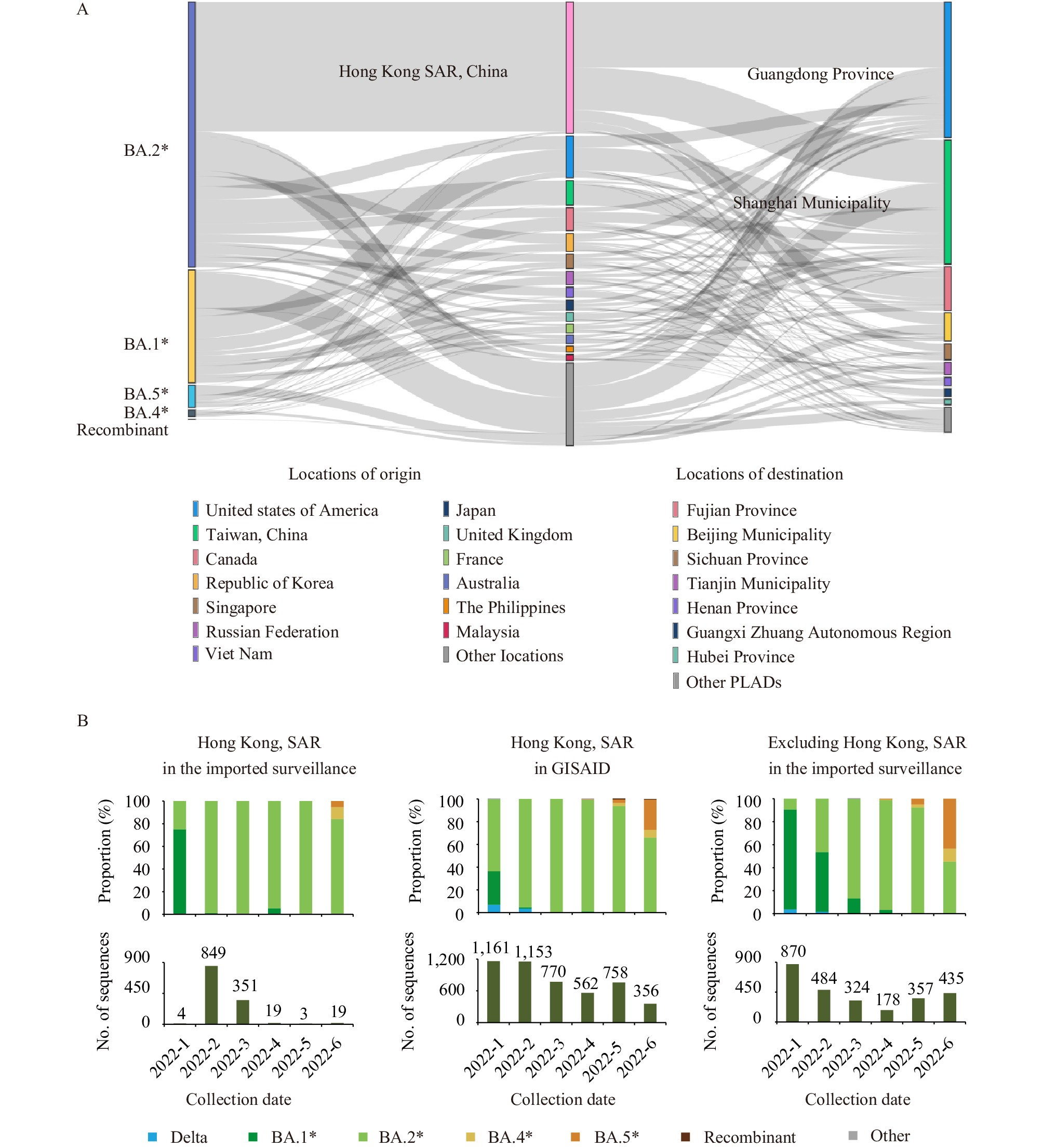 Figure 3.
Figure 3.Specific source analysis of SARS-CoV-2 VOCs. (A) The relationships between the origin and destination of each SARS-CoV-2 Omicron imported case. (B) The proportion of SARS-CoV-2 VOCs from Hong Kong SAR in the imported SARS-CoV-2 surveillance and in GISAID, and the proportion of SARS-CoV-2 VOCs in the imported SARS-CoV-2 surveillance (excluding Hong Kong SAR) from January to June 2022.
Note: In panel A, the columns from left to right were the major lineages of Omicron, location of origin, and location of destination. Other locations: the other 104 locations with a number of sequences less than 50. Other PLADs: the other 15 PLADs with a number of sequences less than 50.
Data from the imported surveillance and GISAID was up to July 31, 2022 and July 17, 2022, respectively.
Abbreviation: SARS-CoV-2=severe acute respiratory syndrome coronavirus 2; VOC=variant of concern; PLAD=provincial-level administrative division; SAR=Special Administrative Region; GISAID=the Global Initiative on Sharing All Influenza Data.
* Includes the descendent lineages.
As Hong Kong SAR had the largest number of cases recorded in the surveillance in the first half of 2022, this investigation compared the data of Hong Kong SAR in GISAID with that in the imported surveillance. In February and March, Hong Kong SAR submitted 1,153 and 770 sequences to GISAID, respectively — among which BA.2 accounted for 95.32% and 100.00%, respectively (Figure 3B). The proportions were similar to that in imported SARS-CoV-2 surveillance (February: 98.94%; March: 99.72%). Other months, except February and March, had fewer sequence records in the imported surveillance. The proportion of SARS-CoV-2 variants in the imported surveillance after the removal of Hong Kong was closer to that of GISAID data in February and March (Figure 2B and Figure 3B).
-
In the present study, genomic sequence surveillance data of SARS-CoV-2 from imported cases in the mainland of China from January to June 2022 were analyzed. The median deposition time in all PLADs submitting sequences was less than 1 month. However, some sequences were submitted 3 months after the sample collection date, which may blunt the surveillance sensitivity. Guangdong and Shanghai had the largest number of submission sequences and covered the widest range of origins, consistent with the results of surveillance data from 2021 (2). It is further suggested that international travel hubs such as Shanghai and Guangdong can be used as surveillance sites to strengthen the monitoring of SARS-CoV-2 global circulation.
From the surveillance data, Omicron dominated in the first half of 2022, shifting from BA.1 to BA.2, then to BA.4 and BA.5. The epidemic trend was in accordance with that in GISAID, while the proportions of each major lineage were quite different, especially in February and March. Based on further analyses of geographic origin, the discrepancy might be partially due to the surge of BA.2 imported cases from Hong Kong. Indeed, since January 2022, Hong Kong has experienced a large-scale COVID-19 epidemic caused by the sub-lineage of BA.2, with a cumulative number of over 1 million reported cases (14). Accordingly, findings from the imported surveillance are likely to be skewed by the case population. Optimizing the directing sequencing efforts toward travelers departing from targeted countries or regions might help overcome this limitation.
Notably, the Omicron recombinant XU imported from the United Arab Emirates was first documented in this imported SARS-CoV-2 surveillance. Only 17 sequences of XU were shared from Japan, India, Australia, the United States of America, the United Kingdom, and Malaysia in GISAID (12). The reasons for the absence of recombinant XU sequences from the United Arab Emirates in GISAID might be associated with genomic surveillance and data-sharing strategies conducted in the United Arab Emirates, indicating that the actual prevalence of some variants may be underestimated. So far, 33 Omicron recombinants have been designated as lineages in the Pango nomenclature system, named XD to XAR (6). With the exception of XE, which has been reported to be 9.8% more transmissible than BA.2 and spread to 28 countries in the first half of 2022 (15-16), the rest of the recombinants, including XU, have not been reported to cause rapid epidemic expansion and little is known about their transmissibility, severity, and neutralization evasion capability. Further surveillance and studies are needed for these recombinants.
There are 3 other limitations to these results. First, the data in GISAID may be affected by the differences in sequencing capacity and sampling strategies among countries, the lag of sequence uploading, or even the decreased number of shared sequences. Second, although PLADs in the mainland of China were required to conduct genome sequencing for all imported COVID-19 cases, the sequenced coverage of imported cases may be affected by sample quality, differences in sequencing capability, and differences in reporting timeliness by PLAD. During the study period, the sequence of imported cases from Xinjiang, Tibet, Qinghai, and Anhui was not received, which may cause bias in lineage proportion and geographic analysis. These aforementioned PLADs should strengthen the sequencing of imported cases and shorten the feedback time of sequences to improve overall national monitoring sensitivity. Third, a lack of information on regions passed through before imported case documentation may mask the true origin of imported cases.
This study’s findings revealed that Omicron has been introduced into the mainland of China multiple times from multiple locations. Far more sequences were recorded in the first half of 2022 than in all of 2021 (2). The surge of imported cases increased the risk of triggering local outbreaks. Since February 2022, the sub-lineage of BA.2 that was circulating in Hong Kong SAR has caused an outbreak in Shanghai with over 600 thousand infections (17). In addition, of the 7 lineages designated as “the Omicron subvariants under monitoring” by WHO, BA.5 was the most dominant strain in June and caused local transmissions in the mainland of China more than once (18). Considering the rapid alternation of the dominant variant within Omicron in the first half of 2022 and the gradual slowing of global genomic sequence surveillance efforts, the long-term genomic surveillance of imported SARS-CoV-2 will continue to play a vital role in the early warning of new variants as well as tracing the source of local transmissions in the mainland of China.
-
No conflicts of interest.
HTML
| Citation: |




 Download:
Download:




