-
Echinococcosis is a zoonotic parasitic disease caused by the Echinococcus larvae (1) that seriously threatens human health and restricts local socioeconomic development. It is widespread in pastoral and semi-pastoral areas in northwestern China, particularly in the Qinghai-Tibet Plateau (2). Humans often become infected by the ingestion of Echinococcus eggs excreted from the feces of a host, such as by having water and food polluted by Echinococcus eggs accidentally, or by touching surrounding soil or dog hair contaminated with Echinococcus eggs (3). However, there is a lack of information so far about other environmental transmission routes for this parasite. Hence, this study aimed to investigate the presence of E. granulosus deoxyribonucleic acid (DNA) in the environment surrounding Daqian Town, Naqu City, Xizang (Tibet) Autonomous Region. The results showed that positive samples were detected via nucleic acid detection method from the environmental samples of residents’ hand-eluent, dog hair, and surrounding soil. Sequencing confirmed that the obtained polymerase chain reaction (PCR) products represented mitochondrial cytochrome c oxidase subunit I (COI) gene fragments from E. granulosus. Thus, this study provides evidence to design targeted interventions as well as optimize existing prevention and control strategies to curb echinococcosis infections.
To understand transmission routes that risk contamination by Echinococcus in the environment surrounding human-habited areas, this study conducted a survey in Xizang (Tibet) Autonomous Region in August 2020. The first action item was the selection of survey sites: through considering the production mode (mainly husbandry) and high levels of echinococcosis in various areas, Daqian Town was resultantly selected (Figure 1). The second was sample collection: a total of 171 samples were collected randomly, including eluent samples from residents’ hands (78 samples), hair samples from dogs (28 samples), topsoil samples from surrounding settings (37 samples), and grass samples from pasture areas (28 samples). The standard collection for samples was as follows: eluent samples from residents’ hands were rinsed with 500 mL water after 1 mL 7X hand sanitizer and filtered through 500 mesh nylon silk cloth; 1 g of dogs’ surface hair was collected; 25 g surface soil within 100 m2 centered on the kennel was collected; and 25 g of surface grass samples were collected. Safety protocols were adhered to during the collection of all samples to avoid infection as infected host feces are the only source of eggs in such an environment.
The third step was nucleic acid detection. Prior to extracting nucleic acid from samples, the samples were processed as follows: soil samples of 15 g were saturated in approximately 100 mL brine; and 200 mL of brine was added to each eluent, hair, and grass sample for the purpose of saturation. Next, samples were placed in a glass vessel on an automatic oscillator (ZWYR-D2403, ZHICHENG, China) for 30 min at 200 rpm and maintained at 25 °C for 40 min (without shaking). In total, 50 mL of supernatant was extracted and filtered by an electric vacuum pump (SHB-III, CHANGCHENG, China) (pore size was 0.22 μm). Extraction of nucleic acids from all samples was performed using the TIANNAMP Soil DNA Kit (No. DP336). After the concentrations of all samples were measured, they were stored at −20 °C for further analysis.
Fourthly, COI gene sequencing: the mitochondrial COI gene of Echinococcus was subjected to PCR analysis. The amplification was performed according to the protocol described by Guo ZH et al (4). The DNA isolated from adult E. granulosus and distilled water were used as positive and negative controls in the PCR experiments, respectively. The final PCR products of the positive samples were sequenced by Sangon Biotech (Shanghai). Sequencing results were compared with the Echinococcus sequence deposited in the GenBank database. Evolutionary analyses were conducted in MEGA 11 software (version 11.0.10, Mega Limited, Auckland, New Zealand) (5). Statistical analysis was performed using the SPSS software package (version 21.0, IBM, Armonk, USA). All data were transferred to Microsoft Excel software (version 2016, Microsoft, Redmond, USA) for data compilation.
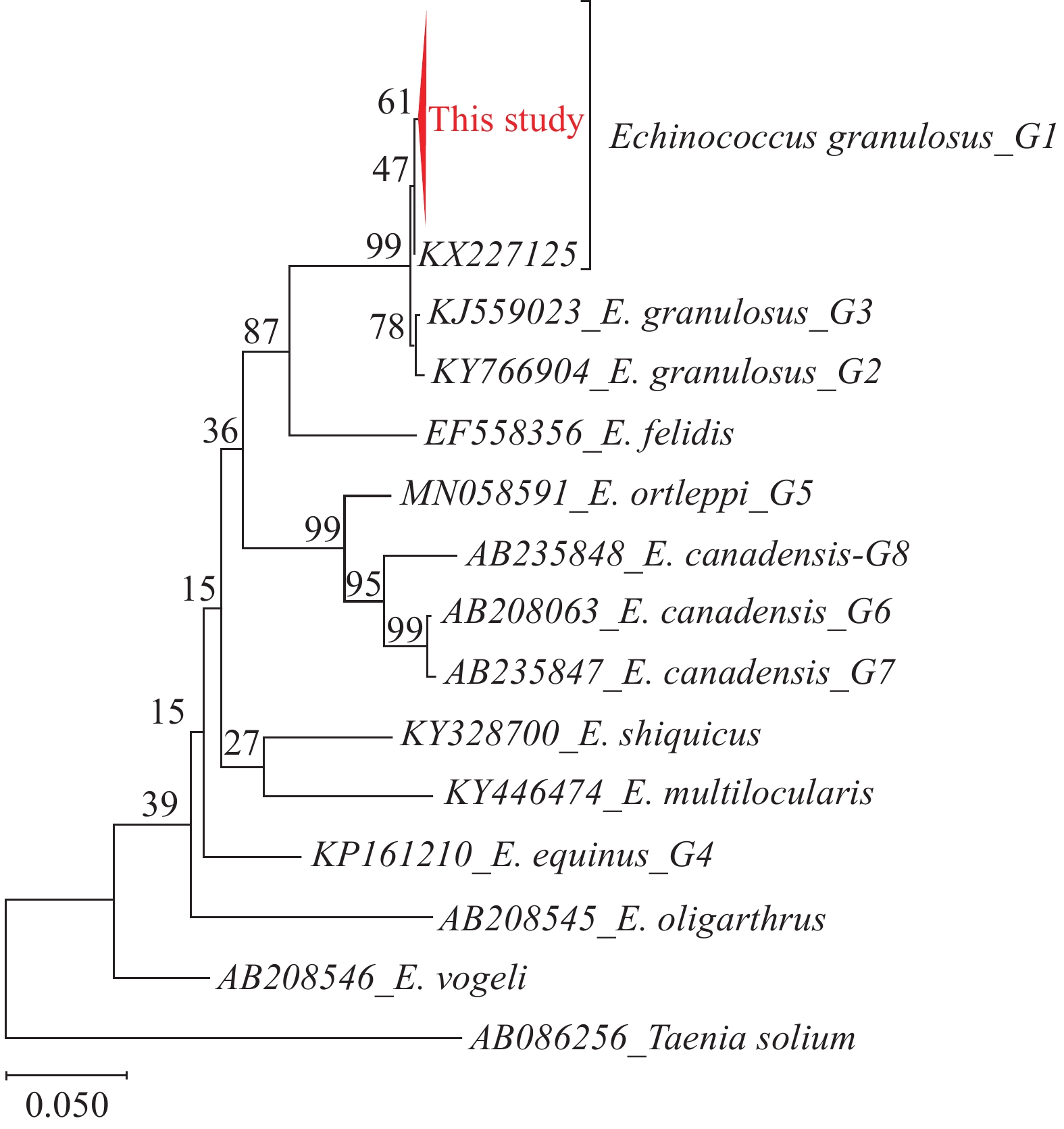
The results showed that the positivity rates of nucleic acid in the eluent samples of residents’ hands, hair samples from dogs, and topsoil samples were 8.97% (7/78), 7.14% (2/28), and 2.70% (1/37), respectively. All grass samples were negative in the detection of nucleic acid (Table 1). The sequencing of selected positive samples and comparison with the Echinococcus sequence deposited in the GenBank database confirmed that the PCR products obtained represented E. granulosus mitochondrial COI gene fragments, and showed 99.19%–100.00% similarity to the referential sequence (accession number: KX227125). Evolutionary analysis showed that the haplotypes may be closer to the G1 type of E. granulosus (Figure 2).
Sample type Number of samples
collected (n)Positive samples Test Number (n) % Eluent from residents’ hands 78 7 8.97 P=0.356 Hair from dogs 28 2 7.14 Soil 37 1 2.70 Grass 28 0 0.00 Total 171 10 5.85 Note: The post-hoc pairwise tests for positivity rates of all sample types were performed, and there were no statistical significances of any pair. P values were adjusted by Bonferroni correction and compared with the 0.05 level. Table 1. 171 environmental sample information and test results, Daqian Town, Naqu City, Xizang (Tibet) Autonomous Region, 2020.
-
Overall, this study investigated the presence of E. granulosus DNA in environments surrounding both where husbandry is practiced and where there is a high rate of echinococcosis cases. This study thus evidences that residents’ hands, dog hair, and topsoil from Daqian Town in Xizang (Tibet) Autonomous Region have been contaminated with this parasite and that local residents may have been infected by their surrounding environment. Notably, the nucleic acid positivity rate of eluent samples from residents’ hands was 8.97% (7/78), indicating that the E. granulosus DNA easily adheres to people’s hands, which is an important transmission route of infection for local residents. The results of nucleotide sequence comparison showed fragments were from E. granulosus, which is consistent with the local prevalence (6). Against this background, understanding the presence of E. granulosus DNA in surrounding settings can help local communities control the transmission risk of echinococcosis more effectively. This study suggests that residents are at risk of catching echinococcosis through hand-oral transmission routes. Health education given to local residents is thus a key intervention with the central message being “wash hands frequently and don’t play with wild dogs.”
Previous studies have shown that Echinococcus eggs may preserve their infectivity for a long time, which could have contributed to the distribution of infectious eggs present in the environment surrounding Daqian Town. The eggs of E. granulosus can be completely inactivated at −80 ℃ for at least 7 days. They can survive for more than 200 days at 7 ℃ and 50 days at 21 ℃ (7). General information regarding the distribution of eggs in the environment is fragmentary. Specifically, very few studies address the contamination of environmental matrices (3,8-10). In Poland, DNA from E. multilocularis eggs was detected in soil collected from forests, arable fields, kitchen gardens and farmyards (9). Similarly, environmental fruit, vegetable, and mushroom samples have also contained the presence of E. multilocularis eggs (10). In this study, samples were tested from dogs’ feces, dog hair, and soil, which were collected in a highly endemic region of Echinococcus in Sichuan Province; the results showed that the positive rates of hair samples from dogs and soil samples were 13.1% (8/61) and 7.7% (3/39) (3). These results show that canine feces will indeed cause contamination to the surrounding environment, and surveillance of DNA from Echinococcus eggs in the environment is necessary. However, there is insufficient information about the occurrence of DNA from Echinococcus eggs on residents’ hands as a result of contact with their environment. As well as providing an indication of hand-oral as a transmission route, this study is the first environmental survey of the presence of E. granulosus DNA from residents’ hands in China. The evolutionary history was inferred using the Maximum Likelihood method and General Time Reversible model. Evolutionary analysis by Maximum Likelihood method in this study showed that the haplotype may be closer to the G1 type of E. granulosus (Figure 2).
However, this study was subject to some limitations. First, due to the large workload of sample collection, the number and variety of samples in this study are small. Our results suggest that more investigations should be performed in the future in order to obtain a clear picture. Second, the study focused on heavy endemic areas and did not involve other areas. We will enlarge the range of sample collection and do further investigations from this aspect in the future.
In conclusion, the results from this study provide scientific reference for the development of echinococcosis prevention and control strategies. As well as providing an indication of some environmental transmission routes of E. granulosus eggs, such a survey is of use in further health education efforts. These results should motivate professional prevention and treatment efforts against echinococcosis, help introduce informative and educational campaigns about the risk of infection, as well as aid in the development of methods for protecting the population — especially those living in at-risk areas.
-
We thank participants of the survey.
-
No conflicts of interest.
HTML
| Citation: |



 Download:
Download: