-
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of coronavirus disease 2019 (COVID-19), is constantly mutating under the different circumstances of global transmission (1). The emerging SARS-CoV-2 variants may have potential adverse impacts on epidemic traits and severity. To some extent, it is also capable of escaping natural and vaccine-induced immunity (2-3). Some of them were designated as variants of concern (VOCs) by the World Health Organization (WHO) (4). Therefore, robust surveillance is essential to assess the evolution of viruses in real time.
After the epidemic in Wuhan City was brought under control in 2020, several COVID-19 outbreaks in the mainland of China have been proven to relate to SARS-CoV-2 contaminated cold-chain products (5-7), while most were caused by transmission through imported cases on flights, at isolation facilities, or in designated hospitals (8-9). Therefore, genomic surveillance for SARS-CoV-2 from imported cases is of great significance for monitoring the risk of different variants that were imported into the mainland of China, assessing the risk of importation-associated domestic spread, and helping guide public health interventions. On March 17, 2020, the China CDC released a notice and launched genomic surveillance for SARS-CoV-2 from imported COVID-19 cases nationwide. The laboratories of provincial CDCs were required to conduct SARS-CoV-2 whole-genome sequencing for samples from imported cases and submit the genomic sequences to the China CDC in time. This study includes the analysis of genomic surveillance data of imported SARS-CoV-2 cases of 2021 from the mainland of China.
-
The sequences of SARS-CoV-2 from laboratories of the provincial CDC were submitted to the China CDC for verification and further analyses. Data from Hong Kong Special Administrative Region (SAR), China, Macao SAR, China, and Taiwan, China were not included in this study. Global SARS-CoV-2 sequence surveillance data were collected from the Global Initiative of Sharing All Influenza Data (GISAID) up to March 9, 2022.
Spatial mapping was conducted to describe the regional distribution of SARS-CoV-2 sequences from imported COVID-19 cases and the timeliness of sequence submission. The standard map [No. GS (2016) 2923] was downloaded without modification from the standard map service website of the National Administration of Surveying, Mapping and Geographic Information. Lineage analysis was conducted on sequences with genomic coverage above 90%. SARS-CoV-2 lineages were assigned using the Phylogenetic Assignment of Named Global Outbreak Lineages web application (PANGOLIN; version 4.0.5) (10). The SARS-CoV-2 VOCs were classified according to the WHO’s designation (4). The proportions of VOCs were calculated based on the date of sample collection. As it takes several days to weeks from sample collection to sequence submission, the number of submitted sequences in December was lower than the actual value.
-
The SARS-CoV-2 genomic sequences of 3,355 imported COVID-19 cases were submitted to the China CDC from January 1 to December 31, 2021 (upon validation, one sequence of them was submitted on December 25, 2020). Except for Xizang (Tibet) Autonomous Region and Qinghai Province, the remaining provincial-level administrative divisions (PLADs) had sequences submitted in various amounts (Figure 1A). The 3 PLADs with the largest submitted sequences were Shanghai Municipality (23.9%, 803/3,355), Yunnan Province (22.9%, 768/3,355), and Guangdong Province (16.9%, 568/3,355), followed by Tianjin Municipality, Fujian Province, Sichuan Province, and Henan Province, which submitted more than 100 sequences each. There were 8 PLADs that submitted 30–100 sequences. The remaining 14 PLADs submitted fewer than 30 sequences each.
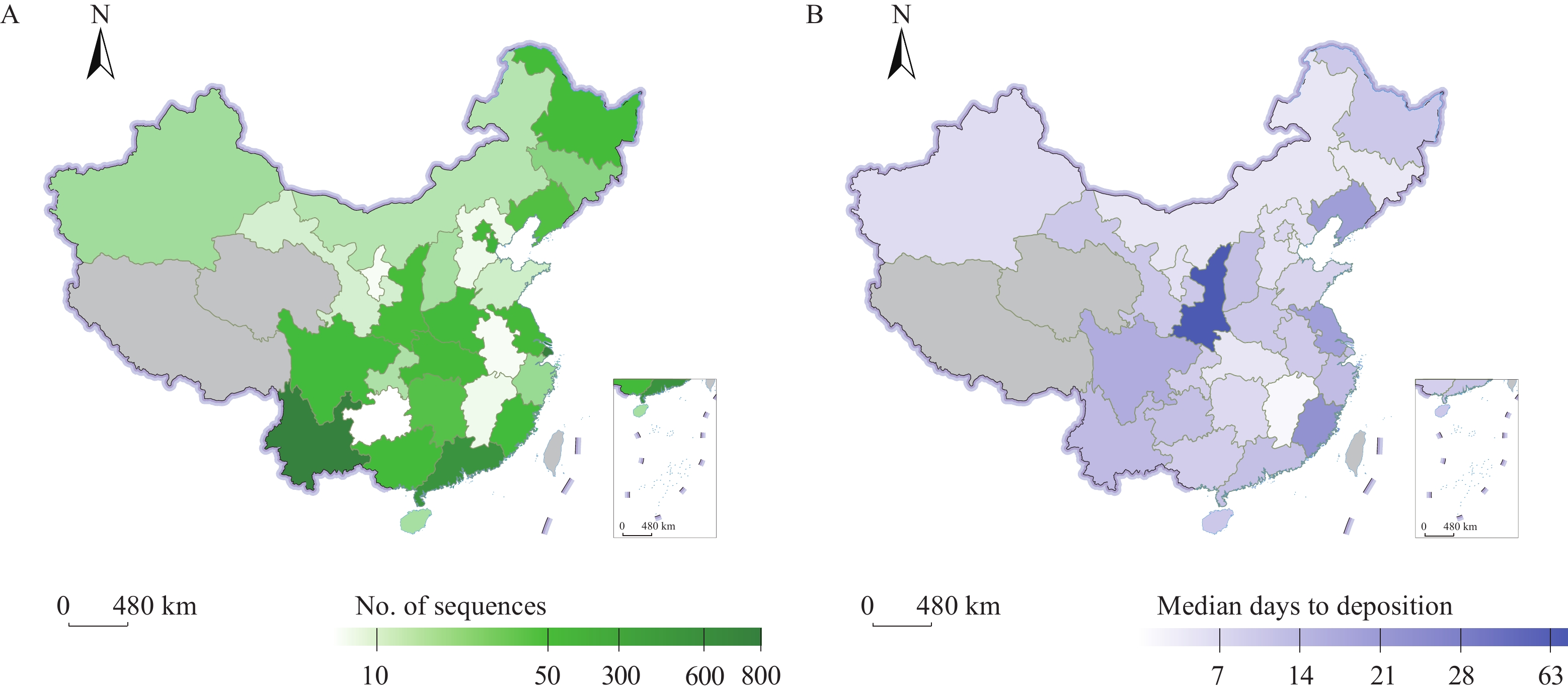 Figure 1.
Figure 1.The surveillance of SARS-CoV-2 genome of imported cases in the mainland of China, 2021. (A) The number of genomic sequences of imported cases submitted by PLADs. (B) The median sequence deposition time of each PLAD from sample collection to sequence submission.
Note: Data as of December 31, 2021. Grey part means no data.
Abbreviation: SARS-CoV-2=severe acute respiratory syndrome coronavirus 2; PLAD=provincial-level administrative division.
To validate the timeliness of the sequence submission, the deposition time of each sequence was calculated according to the date of sample collection and sequence submission. The results showed that Shaanxi Province had the longest median time of sequence deposition with 67 days (Q3-Q1=109-8), followed by Fujian with 23 days (Q3-Q1=44-11). The remaining PLADs submitted sequences within 3 weeks of sample collection (Figure 1B).
Of the 3,355 sequences, 3,309 sequences were collected in 2021. Of the 3,309 sequences, 2,388 (72.2%) had a genomic coverage of >90%, allowing for further lineage analysis (Figure 2A). From January to April, the number of submitted sequences was less than 100 available sequences per month. Since May, the number had increased, with more than 100 available sequences per month. July and August had the highest numbers, with 548 and 407 available sequences, respectively. All five VOCs (Alpha, Beta, Gamma, Delta, and Omicron) were identified in the imported cases. The proportion of the Alpha variant was the highest from February to May. Subsequently, the Delta variant gradually replaced Alpha, becoming dominant in June and overwhelmingly dominant from August to November (more than 95% per month). In December, Omicron variants were detected, accounting for 54.3% (151/278) of the sequences, including BA.1 and BA.2 lineages.
 Figure 2.
Figure 2.The proportion of SARS-CoV-2 VOCs in the imported SARS-CoV-2 surveillance and in GISAID, 2021. (A) The imported SARS-CoV-2 surveillance in the mainland of China (Data as of December 31, 2021). (B) GISAID (Data as of March 19, 2022).
Abbreviation: SARS-CoV-2=severe acute respiratory syndrome coronavirus 2; VOC=variant of concern; GISAID=the Global Initiative of Sharing All Influenza Data.The pattern of prevalence of the VOCs from the imported SARS-CoV-2 surveillance was similar to that in GISAID (Figure 2B). However, the relative proportions of VOCs were different, particularly for the Beta and Gamma variants. From January to May, the proportion of the Beta variant in the imported SARS-CoV-2 surveillance was more than 10% per month, compared to the data of 1.2%–2.2% in GISAID. The proportion of Gamma variant was the highest in April (5.1%, 2/39) and decreased to 1.4% (2/141) and 1.2% (3/259) in May and June, respectively, in the imported SARS-CoV-2 surveillance. In GISAID, the proportion of Gamma variant was 5.9% (26,530/449,653) in April, which increased to 7.5% (25,933/347,752) in May, and reached a maximum of 7.9% (22,660/285,518) in June.
To assess the sensitivity of the surveillance, the time of each VOC for sample collection of the first record, the initial local transmission in the mainland of China as well as its designation by the WHO as a VOC were recorded (Table 1). The time intervals for Alpha, Delta, and Omicron variants between sample collection of their first record in the surveillance and WHO designation were less than one month. Except for Gamma, the time period for the remaining VOCs causing local transmission in the mainland of China was later than that of the sample collection of their first records in the surveillance. There was no local transmission caused by the Gamma variant in the mainland of China.
VOC Earliest recorded sequence in the imported
SARS-CoV-2 surveillanceDate of first local
transmissionEarliest documented
samples in the world†Date of designation
by the WHO†Submitted PLAD Submission date Alpha Shanghai 2020-12-25
(Collection date:
2020-12-14)Beijing,
2021-1-17United Kingdom,
2020-9VOC: 2020-12-18 Beta Hunan 2021-2-7
(Collection date:
2021-1-16)Guangdong,
2021-1-23South Africa,
2020-5VOC: 2020-12-18 Gamma Jiangsu 2021-3-30
(Collection date:
2021-3-18)− Brazil,
2020-11VOC: 2021-1-11 Delta Chongqing 2021-4-28
(Collection date:
2021-4-22)Guangdong,
2021-5-21India,
2020-10VOI: 2021-4-4
VOC: 2021-5-11Omicron (BA.1*) Tianjin 2021-12-13
(Collection date:
2021-12-9)Guangdong,
2021-12-16Multiple countries,
2021-11VUM: 2021-11-24
VOC: 2021-11-26Omicron (BA.2*) Guangdong 2021-12-29
(Collection date:
2021-12-27)Tianjin,
2022-2-26* Includes the descendent lineages.
† According to the official information published by the World Health Organization (4).
Abbreviation: SARS-CoV-2=severe acute respiratory syndrome coronavirus 2; WHO=World Health Organization; VOC=variant of concern; VOI=variant of interest; VUM=variant under monitoring; PLAD=provincial-level administrative division.Table 1. The detection of SARS-CoV-2 VOCs in the mainland of China and the world.
-
This study analyzed SARS-CoV-2 genomic sequence surveillance data from imported cases in the mainland of China in 2021. The 29 provincial CDCs with the submission of sequences performed well overall in terms of the number and timeliness of sequence submissions, but with different characteristics. Notably, the number of sequence submissions is affected by at least two factors. The first was the number of imported cases. In this regard, Shanghai and Guangdong were the main ports of entry for the mainland of China (11), while Yunnan shares a border with Myanmar and has frequent personnel exchanges. Therefore, these PLADs had a high proportion of imported cases. The second was the viral load of the samples. In general, it is difficult to obtain the viral genome from samples with Ct values higher than 35 (12).
The epidemic trend from Alpha to Delta to Omicron in imported cases was consistent with that in GISAID. However, the proportions of Beta and Gamma variants imported to the mainland of China were different from those in GISAID, which may be affected in two ways. First, the data in GISAID may be influenced by differences in sampling strategies, sequencing capacities, and data-sharing time between countries (13). In contrast to the Alpha, Delta, and Omicron variants that were globally prevalent, Beta and Gamma variants were mostly prevalent in Africa and South America, respectively (14). The relatively low number of uploaded sequences in some African countries may explain the low prevalence of Beta variants in GISAID. Second, surveillance data should be interpreted with consideration of the limitations, including the difference in international communication, the number of imported cases, sequence submission, and sequencing capacities between PLADs.
Our data showed that the detection of VOCs in imported cases usually occurred after the WHO designation and before the time of local transmission. Early detection of imported vital variants is helpful for timely adjustment of prevention and control strategies and research. Many countries, including China, have strengthened their genomic surveillance to track the emergence of new VOCs in 2021 (15-16). However, since 2022, several countries have changed their testing strategies, leading to a decrease in the number of sequences shared with GISAID (17). This may have affected the early detection of new VOCs worldwide. The data in this study provides scientific support for strengthening the surveillance of imported SARS-CoV-2 in the mainland of China.
-
No conflicts of interest.
HTML
| Citation: |




 Download:
Download:




