-
On February 6, 2022, an unusual infection event was noticed by hospitals and Beijing CDC: 4 clinically diagnosed typhoid cases (3 in Beijing, 1 in Chifeng City, Inner Mongolia Autonomous Region) were reported to China’s Infectious Disease Information System. The detailed epidemiological investigation was initiated by Changping District CDC. This outbreak involved 23 cases in an apartment in Changping District in Beijing and was caused by extensively drug-resistant (XDR) Salmonella Typhi (S. Typhi) through polluted water supply, which was confirmed by laboratory detection.
-
The index case occurred in a company staff member, female, 24 years old. She was reported on February 2, and her symptoms were high fever (up to 40 ℃ on January 30) and chill. She sought medical service on January 28, polymerase chain reaction (PCR) test for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and antigen test for influenza virus were all negative. S. Typhi was isolated in blood culture. These 4 cases were confirmed by laboratory S. Typhi detection, and all lived in the same rental apartment in the countryside. This apartment had 4 floors, 79 separated rooms, and was self-built by a villager. The villager rented most rooms out, and 106 renters lived in it. The room’s average size was about 10 m2, equipped with a separate bathroom and kitchen.
Since the early 4 reported cases had the same residential history within the typhoid’s incubation period, symptom monitoring was conducted and Salmonella screening by real-time PCR was implemented with stool samples for 106 residents in the apartment and some in a neighboring apartment. As of March 5, 2022, a total of 23 confirmed cases (attack rate 21.7%, 23/106) were identified in this typhoid fever outbreak, and there were no cases from the neighboring apartment. A total of 12 cases were found and tested positive for S. Typhi during the field epidemiological investigation, and the other 11 cases were identified when they sought medical care. All 23 cases developed symptoms, with high fever up to 41 ℃ as the highest (82.6%, 19/23) and diarrhea (47.8%, 11/23). Other symptoms included headache, chills, vomit, nausea, cough, apathy, black stool, and even intestinal bleeding. Their onset date ranged from January 22 to February 13, with the peak on February 4 (Figure 1); 8 cases were male, and the sex ratio was 0.5∶1. All the cases were adults, with an age range between 22 and 45 years old, and median age of 30 years old. They had diverse occupations, but no one was a food practitioner. Even though they lived in the same apartment, cases were scattered on different floors and had no intersection in daily life. All cases were effectively treated with meropenem in designated hospitals according to antibiotic resistance testing result, and most cases were recovered. Close contacts were required to conduct health monitoring, and prophylactic medication could be administered at will. Up to March 5, no new cases have been reported for 20 consecutive days.
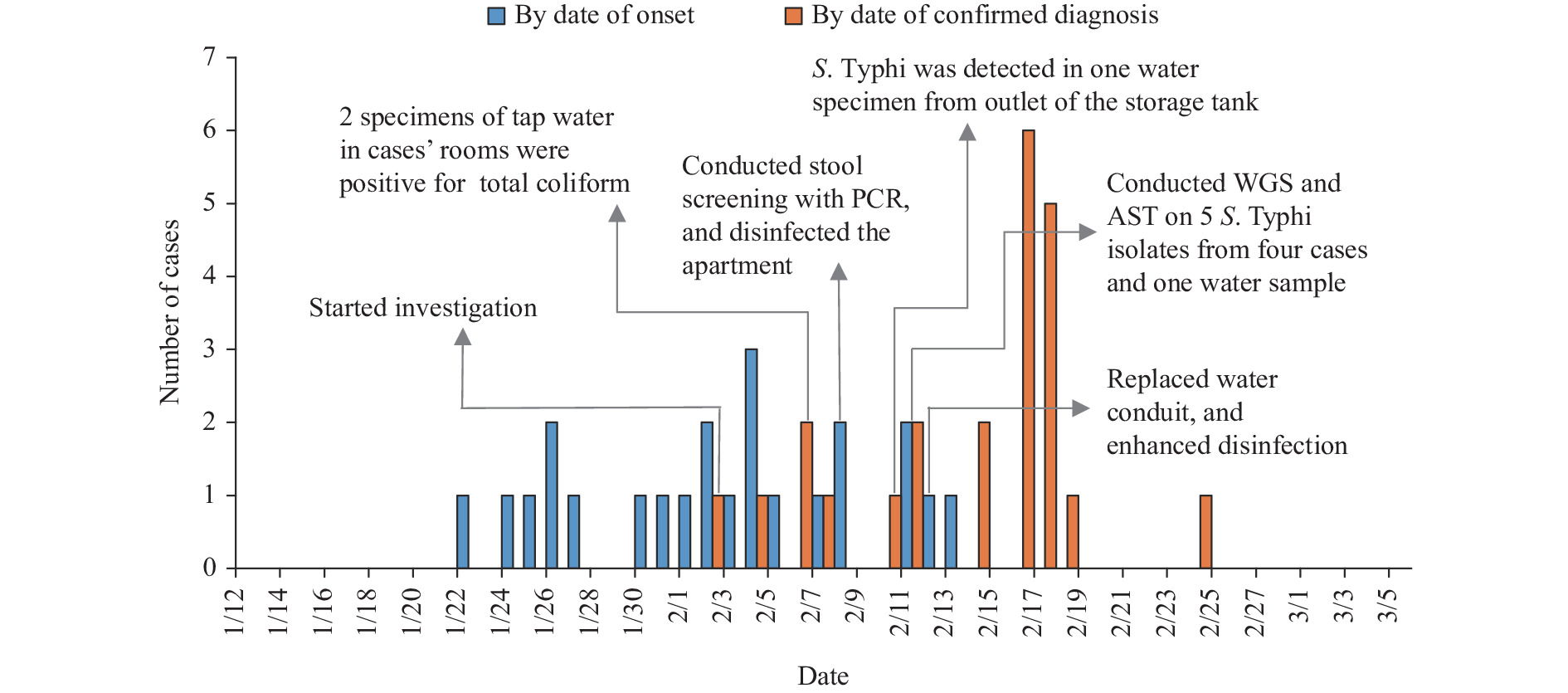 Figure 1.
Figure 1.Epidemiological curve of an XDR S. Typhi outbreak in Beijing municipality, China, January–February 2022.
Note: The epidemiological curve shows the progression of illness in the outbreak over time from the onset of symptoms of the first case on January 22, 2022 through the end of February 13, 2022 (blue pillar), and from the confirmed diagnosis of the first case on February 2, 2022 through the end of February 24, 2022 (red pillar). Key measures and results of the event were marked on the corresponding date. Abbreviations: XDR=extensively drug-resistant; S. Typhi=Salmonella Typhi.Based on the epidemiological curve by date of onset and spatial distribution of cases, a point outbreak was suggested. Considering the possibility of foodborne infection, we checked the dietary history of these cases for 14 days prior to onset, but no evidence of common food or dining together was found to support this. We did not conduct case-control studies, but we received information that most cases keep good hygiene in water usage: they did not drink raw water, but they used tap water to wash vegetables and brush teeth. During this survey, the residents reported that the household water had an odor for about half a month in late December 2021, and some reported that there was disruption of water supply and transient muddy water in mid-January 2022. Herein, the water supply in this apartment was further investigated. The domestic water was supplied from a self-provided well in the village, which was piped to six apartments including the apartment where the cases lived in. Water was pumped from a nearby branch well to storage tank on the roof of the apartment for 24 hours a day to ensure adequate water supply, and then distributed to each room. In field investigation, we learned that sewage pipeline reconstruction work was carried out in December 2021 near the apartment, and the sewage pipeline was just about 1 meter away from the branch well which supplied water for the apartment.
On February 6, we collected 5 water samples (1 from inlet of storage tank, 3 from tap water in 3 cases’ rooms, 1 from tap water in public area), which were tested microbiologically for Escherichia coli (E. coli), total coliform (TC), and Salmonella culture. Results showed very high counts for E. coli and TC in 2 samples of tap water in cases’ rooms, indicating contamination of potable water, but no sample was positive for Salmonella. On February 10, we collected 5 water samples from an inlet of the storage tank again, outlet of storage tank, tap water in cases’ rooms, tap water in unoccupied rooms, and tap water in the neighboring apartment with no cases. One S. Typhi strain was isolated from the outlet of storage tank, while the other four water samples were negative. We collected more water specimens in other apartments and a branch well in the village trying to figure out how this pollution happened, but only some were positive for TC, and S. Typhi was not detected. At the same time, we collected environmental samples including door handles in public areas and case’s rooms, flush buttons, computer keyboards, and mobile phones used by cases, but all were negative for S. Typhi.
Immediate control measures were implemented including health education of the public and emphasis on chlorination of drinking water and sanitation. Then the village committee and the landlord replaced water conduit for the apartment on February 12. We instructed the landlord to clean and disinfect the water tank and water pipeline network, and it was required that the water supply could be restored only after water was eligible according to relevant hygiene standards after two consecutive laboratory tests.
The 5 S. Typhi isolates from the earliest reported 4 cases and 1 water sample were conducted on antibiotic sensitivity test (AST) using the broth microdilution method. All 5 isolates were resistant to ampicillin, chloramphenicol, trimethoprim-sulfamethoxazole, aztreonam, streptomycin, fluoroquinolones, third and fourth-generation cephalosporins, and exhibited extended-spectrum β-lactamase (ESBL) production, but remained sensitive to azithromycin, imipenem, and meropenem, leaving limited treatment options.
Whole-genome sequencing (WGS) of these 5 isolates was carried out using a combination of Nanopore and Illumina Miseq sequencing methods. The complete reconstruction of the entire genome included the final assembled chromosome of 4,896,933 bp in length along with a plasmid of 84,492 bp. The WGS’s results confirmed that all isolates were sequence type ST1 and identified as H58 lineage 4.3.1.1.P1 under the Genotyphi scheme (1-2), obviously different from the previous local isolates, and the same as the strains with extensive drug resistance (XDR) from the 2016 outbreak reported in Sindh Province, Pakistan (3-4). S. Typhi resistant to five kinds of antibiotics (i.e., chloramphenicol, ampicillin, co-trimoxazole, fluoroquinolones, and third-generation cephalosporins), was referred to as “extensively drug-resistant” (XDR) (4-5). These 5 XDR S. Typhi isolates carried an IncY plasmid with the same sequence, harboring a gene cluster of Type 4 secretion system and 6 resistance elements including sul2, aph(3'')-Ib, aph (6) -Id, blaTEM-1B, blaCTX-M-15, and as well as the qnrS fluoroquinolone resistance gene (Figure 2). The genome sequences of 5 strains were submitted into the databank of Chinese Pathogen Identification Net for comparison analysis, which showed that these five strains clustered together, with a clear distinction from other strains.
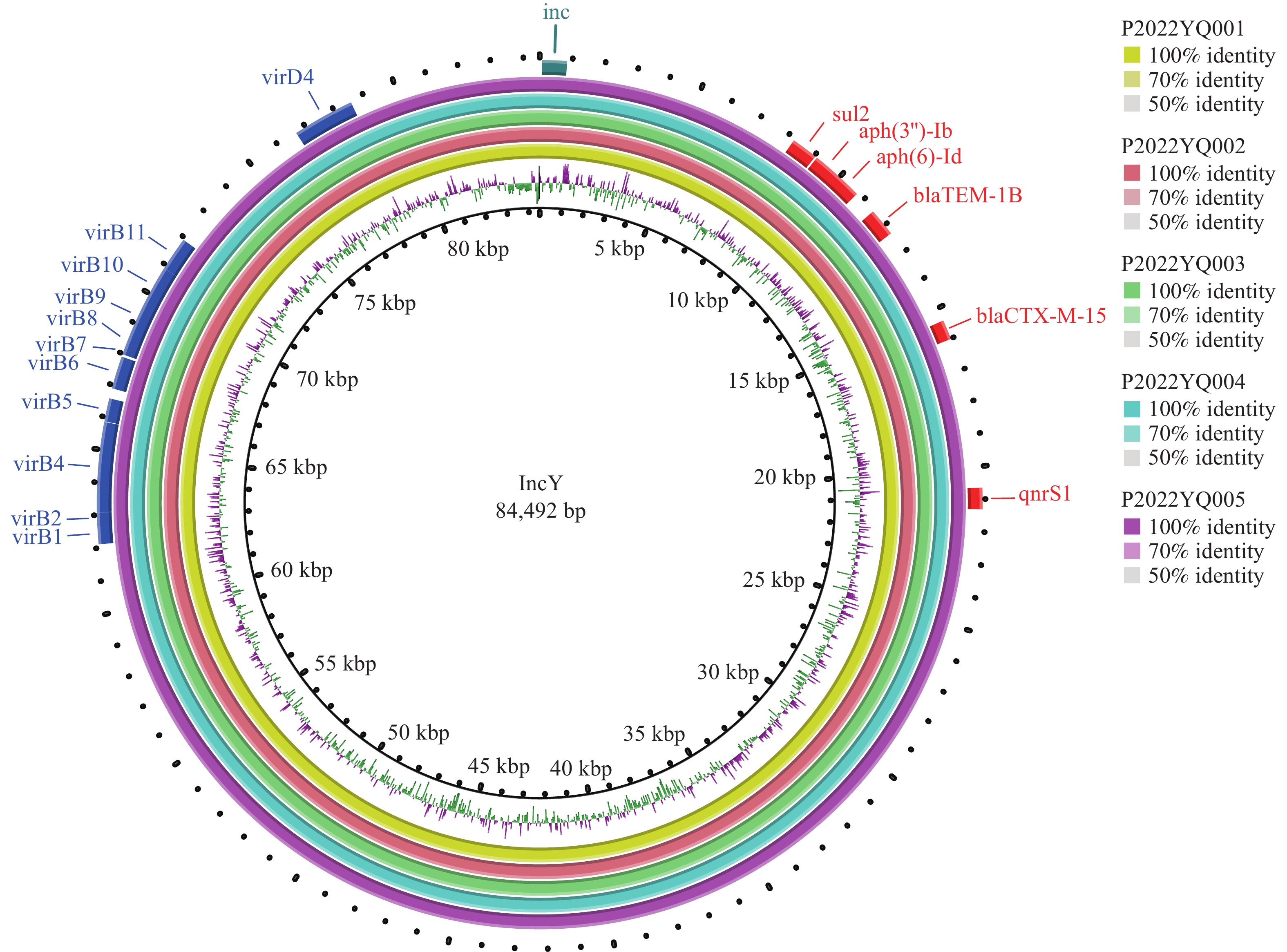 Figure 2.
Figure 2.The plasmid defined as IncY from five XDR S. Typhi isolates.
Notes: The plasmid sequences were annotated using the online tool RAST (http://rast.nmpdr.org/). ResFinder and PlasmidFinder (http://cge.cbs.dtu.dk/services/) were used to identify antimicrobial resistance genes and plasmid replicon types. The plasmids comparison maps were constructed and displayed by using BRIG. Abbreviations: XDR=extensively drug-resistant; S. Typhi=Salmonella Typhi. -
Based on the clinical features and epidemiological characteristics of the cases, evidence of field sanitary survey, and laboratory analysis, we concluded that this outbreak was caused by consuming contaminated water, which was the first report of a waterborne outbreak caused by XDR S. Typhi in Beijing, and to our knowledge, there were no XDR S. Typhi outbreak reports in China. The water supply pollution by S. Typhi might be related to sewage leakage.
Many domestic and international studies had reported that outbreaks of typhoid fever were related to water supply contamination or sewage pipeline leakages. In the typhoid outbreak of Shache City of Xinjiang Uygur Autonomous Region during July-October 2010, epidemiological studies revealed that the consumption of raw tap water from the main water tower of the town was closely associated with the outbreak (6). In Kampala, Uganda, January-June 2015, contaminated water and street-vended beverages were thought to be likely vehicles of this large-scale and persistent outbreaks of typhoid fever (7). More recently, the World Health Organization (WHO) recorded that there were 5,274 cases of XDR typhoid fever out of a total of 8,188 cases of typhoid fever reported in Pakistan from November 2016 up to December 2018 (4). In the globally largest typhoid outbreak survey, the probability of contaminated water as a potential source of XDR S. Typhi was high. Household water cultures conducted had a continuously high E. coli count indicating sewage contamination, but none were positive for S. Typhi. In addition, geospatial mapping revealed clustering of the cases around sewage lines (3).
The above studies lacked direct laboratory evidence to confirm the infection source of typhoid fever outbreak, generally based on epidemiological investigation, case-control analysis, or microbial detection indicators, which indicated that it was not easy to isolate S. Typhi from water samples. Bacteria might survive in the environment in some special forms. Viable but non-culturable (VBNC) bacteria have attracted widespread attention since they were inherently undetected by traditional culture-dependent or molecular methods in the VBNC state. Importantly, VBNC bacteria could resuscitate under favorable conditions leading to significant public health concerns (8). Waterborne pathogens such as E. coli, Salmonella enterica, and Vibrio cholerae could also form biofilms grown in drinking water distribution system (9). These special forms of bacteria decreased the success of culture.
In summary, this was the first report of waterborne outbreak caused by XDR S. Typhi in China. Whole-genome comparison and drug resistance analysis indicated that it belonged to H58 lineage 4.3.1.1.P1 originating from Pakistan, which had the capacity to invade and spread globally by travel-associated international transmission, with the potential to replace native strains (4). However, it was unclear how this novel clone strain entered China and was associated with this outbreak, due to the lack of detailed historical epidemiological data. Importantly, in a modern city such as Beijing, especially in suburban or rural areas where the municipal water supply does not reach, there is still a potential risk of typhoid fever outbreak. So, it is urgent to appeal to relevant governmental authorities to provide safe and hygienic potable water, strengthen supervision on water quality, and educate the public to keep good hygiene habits. In addition, with narrow treatment options for typhoid fever, XDR typhoid itself should also attract great attention, which may lead to treatment failure, prolonged hospitalization, as well as recurrent and extensive transmission of the disease. Therefore, it is necessary to track the source of the XDR strains and to strengthen monitoring their spread through laboratory and extensive epidemiological investigations in the future.
-
Staff of relevant hospital involved in clinical diagnosis and treatment.
HTML
| Citation: |



 Download:
Download:




