-
On January 15, 2022, a 26 years old female (Case A) went to a sampling site for coronavirus disease 2019 (COVID-19) testing due to excessive fatigue and fever for two days. This patient preliminarily tested positive for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using a quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) method. Beijing CDC subsequently verified the result. Furthermore, a rapid site mutation test based on qRT-PCR showed that the virus carried the Q498R mutations but lacked the L452R, T478K, and P681R mutations (ABT, Beijing, China and Bojie), indicating the case was infected by the Omicron variant.
In the following seven days, five close contacts tested positive in screenings, including her mother (Case B), a colleague of the index case (Case C), and three family members of Case C (Case D, E, and F).
The field investigation was performed to identify the source of infection. Case A lived and worked in Haidian District, Beijing, without travel history out of Beijing or close contact with local or imported high-risk populations. Further investigation indicated that the case sent and received international mail occasionally at no fixed interval, due to the need of her work. Notably, an internationally mailed document was received by the case on January 11 (2 days before onset of symptoms), which was delivered from abroad on January 7. The disinfection was carried out from the outer surface of the packaged document when it arrived in Beijing. A total of 1,054 environmental samples were collected, including 22 swabs for the package received by the case. The qRT-PCR showed that 12 of 22 samples were positive in the SARS-CoV-2 ORF1ab/N gene test, including 2 samples collected from the outer surface of the package, 2 from the inner package, and the other 8 from contained papers. Moreover, the rapid site mutation test suggested all 12 positive samples carried the Q498R mutation, which inferred infection with the Omicron variant.
The respiratory samples from all 6 cases and 12 environmental samples, including the inner package and contained papers that were untouched by the case, were used for next generation sequencing (NGS). The viral RNA was reverse transcribed and amplified using ULSEN® 2019-nCoV Whole-Genome Capture Kit (Beijing MicroFuture Technology Co., Ltd, Beijing, China). Then, the sequencing libraries were prepared using the Illumina Nextera® XT Library Prep Kit (Illumina, Inc., San Diego, CA, USA) and were conducted on Illumina MiniSeq platform. Six full genomes were collected from the cases. Overall, 1 full genome and 1 near-full-length genome with a coverage of 88.8% were obtained from environmental samples due to the low viral load. The sequencing analysis concluded that the whole virus belonged to lineage BA.1, Variants of Concern (VOC)/Omicron. In addition, 7 full genomes and 1 near-full-length genome shared a nucleotide similarity of 100%. Phylogenetic analysis indicated the virus located near the cluster of some strains collected in North American and Southeast Asia in mid-December, which had significant differences with local clusters in China in the same period (Figure 1).
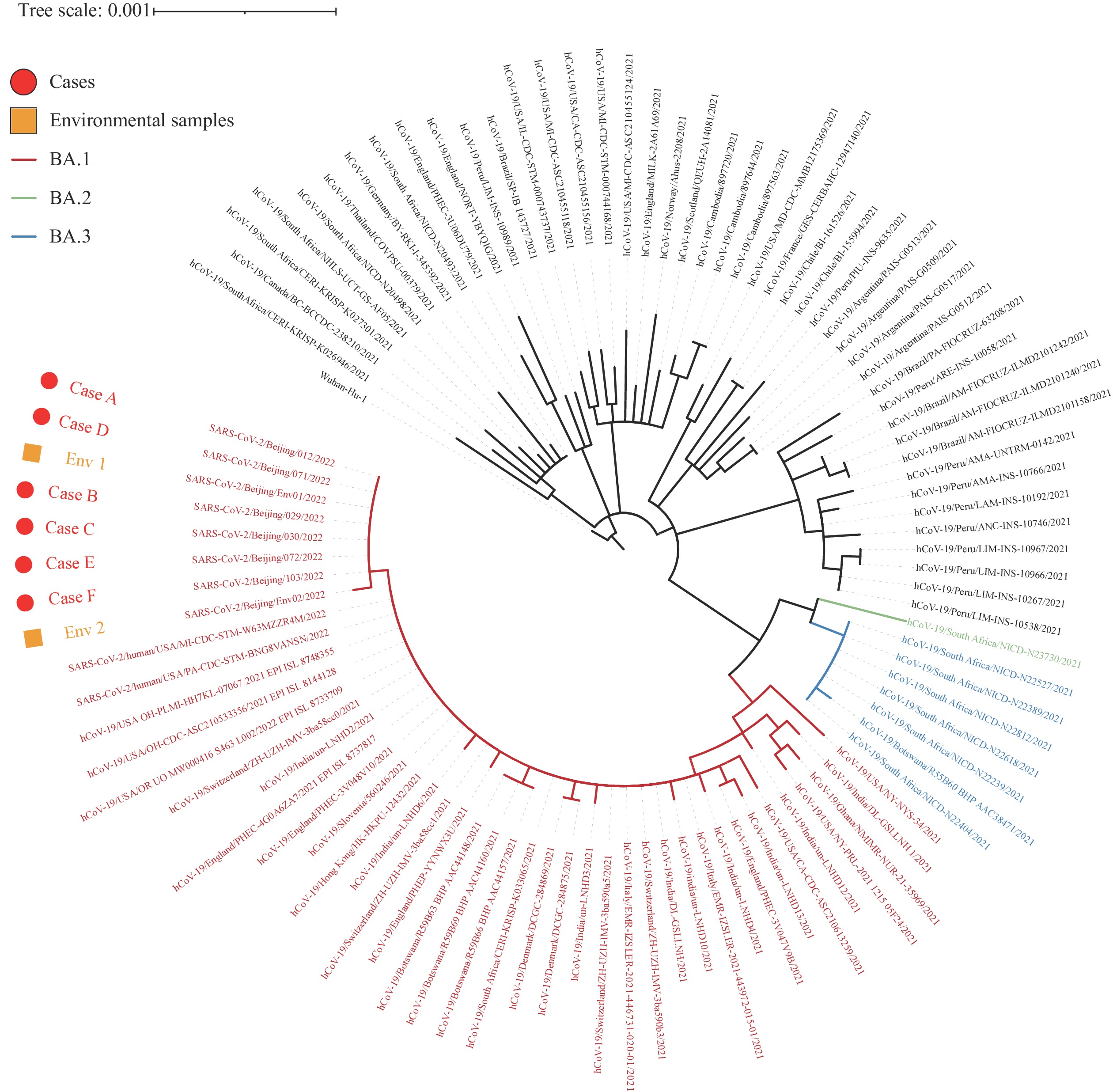 Figure 1.
Figure 1.Neighbor-joining phylogenetic tree for the local cluster of Omicron Variant in Beijing, China, 2022.
Note: The six genomes from Case A to E were indicated by dots. The two genomes from environmental samples were indicated by squares. The major (VOC)/Omicron lineages were colored. The tree was rooted using strain Wuhan-Hu-1 (EPI_ISL_402125).Abbreviations: VOC=variants of concern; Env 1=environmental sample 1; Env 2=environmental sample 2.This is the first local cluster caused by Omicron variant in Beijing. The Omicron variant was first reported on November 9, 2021 and was listed as a VOC by the World Health Organization (WHO) (1-2). As of February 1, 2022, this variant has been identified in more than 170 countries and has become the dominant strain worldwide (3). A remarkable finding of this study was the possible source of the infection for this cluster. We proposed the infection was induced by the internationally mailed document, mainly based on the following evidence. Field investigations showed no potential exposure of the case except the internationally mailed document, with an onset interval of 2 days. All identified cases showed epidemiological links with Case A. Environmental surveillance identified SARS-CoV-2 RNA positive samples from the package’s contents, and parts of these positive contents were untouched by the case. More importantly, NGS showed that the genome of the case matched the samples collected from the mailed documents, which differed from other local strains in China.
Previous studies investigated the virus survival under various conditions. Chin et al. showed SARS-CoV-2 could only survive on printed papers and tissue papers for 3 hours; while on treated smooth papers (such as banknote), it could survive for 4 days (4). Van Doremalen et al. found that no viable SARS-CoV-2 could be measured after 24 hours on cardboard (5). In general, SARS-CoV-2 was less stable on papers than on plastic and stainless steel. However, the temperature should not be neglected during the assessment for viral survival. The low temperatures in winter and in some countries located at high latitudes might lead to an extended viral survival period (4) and should be considered as an essential factor in this cluster.
In summary, we reported the first local cluster caused by Omicron variant in Beijing and showed this cluster was likely caused by a contaminated internationally mailed document, which has not been reported before. Our data emphasized the surveillance and disinfection of imported express cargo in COVID-19 control, especially in certain seasons and regions in China.
HTML
-
All contributing health workers.
| Citation: |



 Download:
Download:




