-
The coronavirus disease 2019 (COVID-19) pandemic was caused by a novel type of coronavirus also known as SARS-CoV-2, 2019-nCoV, and HCoV-19 (1) and quickly spread around the world becoming a major threat to public health (2-3). Although COVID-19 virus is characterized as possessing a large genome and having limited genetic diversity (4) like other coronaviruses due to the high fidelity of its replication mechanism, many single nucleotide polymorphisms (SNPs) have been detected so far. Among the reported SNPs, the strain with the G614 mutation in the S protein has replaced the strain containing D614 as the world’s primary pandemic strain (5). However, it was still unclear if COVID-19 virus strains with G614 were more transmissible than those with D614 (6).
We reconstructed several transmission chains of COVID-19 during the early epidemic phase for 3 countries (Australia, the UK, and USA) based on genomic data from GISAID (7) and Bayesian inference under an epidemiological model for strains with D614 and G614 due to the similar amount of genomes within each country (Figure 1A). Then we inferred the R0 in those transmission chains to compare the difference of transmissibility among humans between D614 and G614 (see the Supplementary Material for details). The R0 caused by G614 was significantly higher than that caused by D614 in Australia and USA, as there was no intersection of 95% confidence intervals (CI) of the 2 distributions of R0 (Figure 1B). For the UK, the mean value of R0 for G614 was slightly higher than D614. For G614, its mean value of R0 was outside the 95% CI of the estimated R0 of D614. The mean value of R0 of D614 also was outside the 95% CI of the estimated R0 of G614. In addition, the R0 for D614 was similar among three countries (from 1.56 for USA to 1.73 for Australia). However, significantly different R0 was detected among countries (from 1.82 for the UK to 3.87 for USA), indicating that the spatial transmission of G614 had higher variation than D614.
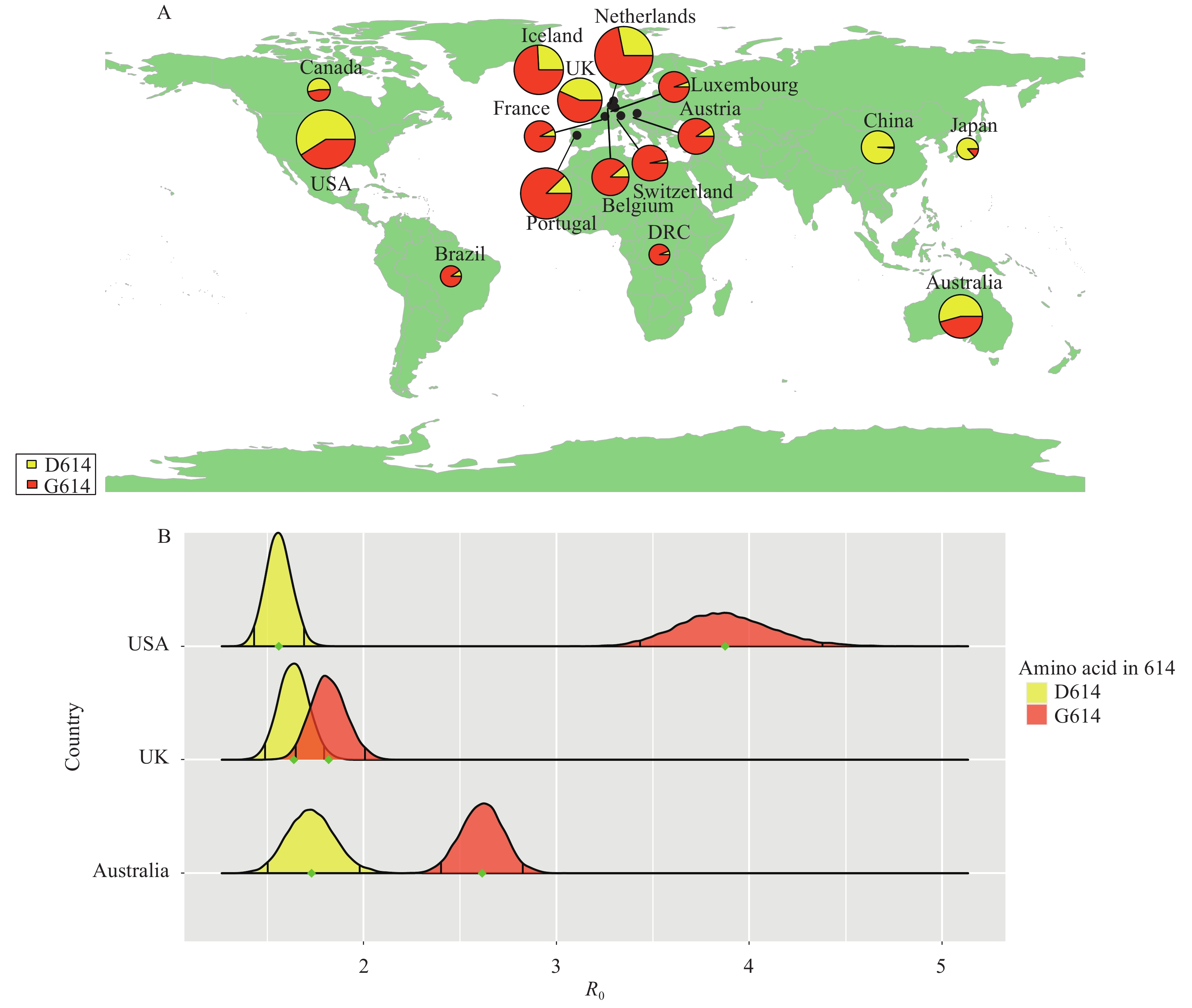 Figure 1.
Figure 1.The statistics and transmissibility of the COVID-19 virus with D614 and G614 during the early phases of the pandemic within each country.
(A) The number of publicly available genomes with D614 and G614 during the first two months after the first confirmed case in each country. The size of the pie chart is associated with the number of genomes. (B) The distribution of R0 for D614 and G614 during the first two months after the first confirmed case in each country. The mean of R0 is represented by a green point. The black line within the distribution represents the 95% confidence interval (CI).Since the data used in this study were all collected during the early phases of each countries’ outbreak and no stringent prevention and control strategies were implemented in these three countries in those phases, the estimation of R0 would not be affected by non-pharmacological intervention. Thus, the result would better reflect the true nature of transmissibility within humans for strains with different mutations. Our findings demonstrated that the G614 mutation accelerated the transmission of the COVID-19 virus and also had higher spatial transmissibility, indicating that strains with G614, which were the dominant strains around the world, could spread on a larger scale and be more difficult to control. These results also echo those experimental results in vitro, in which both clinical samples and pseudoviruses with G614 have higher levels of viral RNA and titers compared to those with D614 (5).
As the COVID-19 pandemic spreads and continues, real-time monitoring and evaluation of the impacts of COVID-19 virus strain mutations need to be consistently maintained to provide earlier warnings for the public and provide evidence that supports government-led countermeasures and strategies.
Acknowledgments: We gratefully acknowledge the authors from the originating laboratories and the submitting laboratories where genetic sequence data were generated and shared via GISAID, enabling this research; Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB29010202, XDB29010102 and XDA19090118); National Science and Technology Major Project (Grant No. 2018ZX10733403, and 2018ZX10713001-010); National Natural Science Foundation of China (NSFC) (Grant No. 32041010 and 31900155); NSFC Outstanding Young Scholars (Grant No. 31822055); and Youth Innovation Promotion Association of CAS (Grant No. 2017122).
HTML
| Citation: |




 Download:
Download:




