-
Introduction: Determining the transmission chain of a virus in its incipient stages is extremely time consuming in traditional approaches that rely mainly on case incidence and interview-based contact data. With the development of high-throughput sequencing technology, genome-based epidemiology approach is showing promise in detecting viral transmission. However, there is still insufficient evidence for the relationship between the viral genetic variations and real viral transmission.
Methods: To explore the possible relationship between transmission chains and viral genetic variations, we combined both epidemiological data and viral genomes of COVID-19 virus collected from Sichuan Province. A phylogenetic approach was used to infer the transmission chain, which was then compared to the transmission chain that came from epidemiological data.
Results: We found that the putative transmission chains were highly concordant to the true transmission chains from epidemiological data, suggesting a strong correlation between viral genetic variations and the viral transmission chain.
Discussion: Our results showed advantages of viral genomic sequencing in tracking and perceiving pathogen transmission, which allowed for potential improvements in the design and implementation of population-level public health interventions.
-
It is crucial to understand the evolution and transmission of a virus in its incipient pandemic, especially the transmission chains, which can help to design effective strategies for disease control and prevention (1). However, it is time consuming and costly to ascertain transmission chains traditionally, in which the process largely relies on case incidence data and interview-based contact tracing (2). A recent study on the epidemiology of coronavirus disease 2019 (COVID-19) in Guangdong Province, China found the transmissions from epidemiological data were congruent with the phylogenetic cluster patterns that inferred from single nucleotide variations (SNVs) of COVID-19 virus genomes (3), suggesting the possibility to infer transmission chains from viral genomes. To investigate possible correlations between transmission chains and viral genetic variations, we combined both epidemiological data and viral genomes of COVID-19 virus collected from Sichuan Province. We found the putative transmission chains are highly concordant to the true transmission chains from epidemiological data. Given to the low cost and convenience of sequencing viral genomes, this approach showing promise to assist disease control and prevention during viral pandemic.
-
A total of 44 samples were collected from 44 patients, who tested positive with COVID-19 infection by real-time reverse transcription polymerase chain reaction (RT-PCR). These samples can be further divided from two different batches. Batch 1 includes 29 local patient samples in Sichuan Province collected in early February, whereas batch 2 includes samples from 14 asymptomatic imported COVID-19 patients in mid-March, all returning to China from Egypt. The clinical and epidemiological data were originally collected by multiple hospitals in Sichuan Province and centralized by Sichuan Provincial CDC.
-
An untargeted metagenomic sequencing approach was used to detect and acquire the COVID-19 virus genomic sequences. RNA-based metagenomic sequencing libraries were prepared according to the manufacturer’s protocol of KAPA Stranded RNA-Seq Library Preparation Kit (Illumina Platforms). Metagenomic sequencing was performed on Illumina Novaseq 6000 sequencing platform that hosted by HitGen Inc.
-
Raw reads were further processed by Trim Galore (V0.6.4_dev) for adaptors removal and quality control. Clean reads were mapped to the reference genome of COVID-19 virus (NCBI reference sequence: NC_045512.2) using Bowtie2 (V2.4.1, University of Maryland). Variants were called using LoFreq (V2, Genome Institute of Singapore) with default parameters and filtered to only keep SNVs belong to primary strain (SNVs with allele frequencies ≥0.5) for each individual. To make sure the multiple sequence alignments were in the length, consensus sequences for these individuals were reconstructed from the filtered VCF files using the consensus function incorporated in BCFtools (V1.9, Wellcome Sanger Institute).
Multiple sequence alignment was performed using MAFFT (V7.4, Osaka University). Following alignment, we used maximum likelihood (ML) tree, neighbor-joining (NJ) tree, and Bayesian coalescent tree to explore the phylogenetic structure of these COVID-19 virus strains. FastTree (V2.1.10, Lawrence Berkeley National Laboratory) and BEAST (V2, The University of Auckland) were used to construct the ML tree and Bayesian coalescent tree, respectively. The NJ tree was built using the ape (V5.3) package in R (V 3.6.1, R Foundation). The ML and NJ tree were visualized using FigTree (V1.4.4, University of Edinburgh). The Bayesian coalescent trees were visualized using DensiTree (V2.2.6, The University of Auckland).
-
The genome coverage of COVID-19 virus from 29 local patients’ samples are all above 98%, with average sequencing depth ranging from 23 to more than 11,400. For 14 imported patients, only nCoVTS0414-6 sample has 90.13% virus genome coverage with an average sequencing depth of 6.07. The rest of the samples have more than 98% genome coverage with average depths from 15 to 13,241.
A total of 6 transmission chains were found among batch 1 COVID-19 samples according to the epidemiological data. Surprisingly, we found COVID-19 virus strains that belong to the same transmission chain were tightly clustered on Bayesian coalescent tree (Figure 1), in spite of sporadic dispersions. Notably, even though COVID-19 virus strains from the same transmission chain were clustered together, we found difficulty distinguishing the introduced case from the other COVID-19 cases within each transmission chain. Collectively, using the heterogeneous data apart from Guangdong Province, we showed the phylogenetic results from genomic data were highly concordant to the transmission chain from epidemiological data.
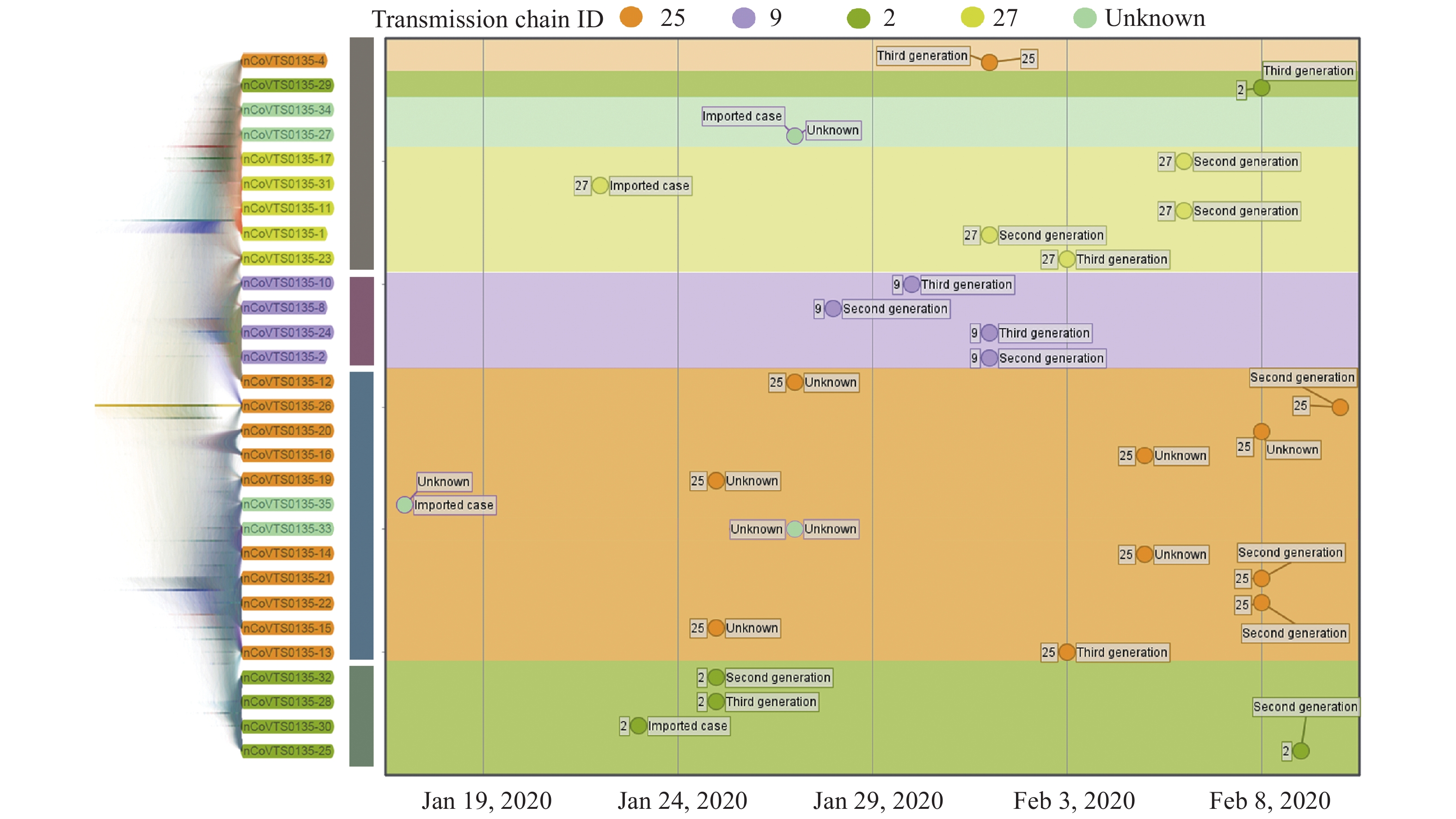 Figure 1.
Figure 1.Phylogenetic relationship of COVID-19 virus strains and the transmission chains from 29 samples. Each dot in the figure represents a patient sample. The onset date of each patient was plotted on the x-axis. The y-axis of these samples were organized based on the position from the Bayesian coalescent tree result. The transmission chain and the generation of the sample in the chain were labelled on the left and right side of each dot, respectively. It was evident that the preliminary transmission chains could be constructed by combining the onset date and the phylogenetic relationship of the COVID-19 virus strains even with limited epidemiological information.
To explore the optimal phylogenetic methods for this study, we also compared the topologies of Bayesian coalescent tree with ML tree and NJ tree (Figure 2). We found the clustering pattern between Bayesian coalescent tree and ML tree were similar to each other and were highly concordant to the transmission chains reconstructed from epidemiological data. However, the clustering pattern of the NJ tree was distinct from the Bayesian coalescent tree and the ML tree, as well as the reconstructed transmission chains. Our results indicated that the NJ tree was incapable of correlating the genomic variations with the transmission chain as accurately as the Bayesian coalescent tree and the ML tree.
 Figure 2.
Figure 2.The phylogenetic analyses showed distinct structures between maximum likelihood (ML) tree and neighbor-joining (NJ) tree. (A) The phylogenetic tree was constructed using maximum likelihood method. (B) The phylogenetic tree was constructed using neighbor-joining method.
Among positive COVID-19 cases, a considerable proportion were asymptomatic (4), which brought significant challenges to epidemic prevention and control because of the difficulty of getting transmission information from the epidemiological data (5). Mutations accumulated in the virus genome during host-to-host transmission could potentially provide more information for transmission chain construction (6). To verify this hypothesis, we conducted phylogenetic analysis using 14 asymptomatic COVID-19 cases. Interestingly, we found that clustering patterns of 14 asymptomatic cases on the Bayesian coalescent tree were highly correlated with travel history of patients (Figure 3A). Among which, patients with COVID-19 virus strains clustered into clade A and clade B had European travel history, while those who clustered into clade C and clade D had Pacific Rim travel history. Moreover, we found the clustering pattern was largely affected by the G28883C mutation, which is a missense variant that lead to a p.204G>R change on QHD43423.2 (Figure 3A). We further found this mutation had a much higher frequency in European COVID-19 cases compared to other regions (Figure 3B), indicating COVID-19 virus strains carrying this mutation might have originated in Europe. Taken together, our results suggested that the Bayesian coalescent method can be helpful in inferring the transmission relationship of asymptomatic COVID-19 cases.
 Figure 3.
Figure 3.Phylogenetic analyses on 14 asymptomatic cases revealed high correlation between travel history and viral phylogenies. (A) The distribution of COVID-19 virus variations among 14 asymptomatic COVID-19 cases and the variations underlie phylogenetic structure. The left panel showed the Bayesian coalescent tree, and the right panel showed the variations in each COVID-19 virus strain. (B) The bar plot shows the regional distribution of G28883C mutation.
-
Targeted interventions have been proven to be an effective way for constraining timing, size, and duration of the epidemic of COVID-19, especially in the early phases of an outbreak (3,7). However, the design of targeted interventions depends on rapid recognizing of an outbreak, which are rarely achieved by traditional approaches through clinical and epidemiological data, especially for asymptomatic infections. Fortunately, with the development of high-throughput sequencing, using pathogen genomes to understand pathogen transmission appears to be possible. In this study, we explored the possibility of inferring transmission chains from genomic sequences using epidemiological data of COVID-19 virus in Sichuan Province. We found that the transmission relationships inferred from genomic variations of COVID-19 virus were highly concordant to the transmission chains that were reconstructed from clinical and epidemiological data. This finding is consistent with the epidemiological study of COVID-19 virus in Guangdong Province (3) and the epidemiology study of Zika virus in the United States (8). However, we were not able to get similar phylogenetic structures among the Bayesian coalescent tree, the ML tree, and the NJ tree. This difference might be caused by the low genetic diversity among COVID-19 virus strains in our study due to regional sampling. As the Bayesian coalescent method is based on aggregate numbers of mutations, which is more robust compared to other methods when there are limited variations within populations (9).
Asymptomatic cases are difficult to recognize without nucleic acid amplification tests. Thus, it is nearly impossible to reconstruct the transmission chains for asymptomatic cases due to their limited clinical and epidemiological data. Using the genomic epidemiology approach in this study, we inferred the transmission relationships among 14 asymptomatic cases and found their phylogenetic relationships were highly correlated with each patient’s travel history. Our results suggested advantages of genomic epidemiology in surveying the spread of asymptomatic cases.
This study was still subject to some limitations. Despite the positive results, this study was still limited by regional sampling. As mentioned by a previous study, underdamping of regions with high incidence can bias phylogenetic analyses (3). Nevertheless, our results showed advantages in the speed and granularity of viral genomic sequencing in tracking and perceiving pathogen transmission, which allowed for potential improvements in the design and implementation of population-level public health interventions.
Acknowledgments: Sichuan Science and Technology Program (nos. 2020FYS0015 and 2020FYS0017); and the National Natural Science Foundation of China (no. 82041033).
HTML
Clinical and Epidemiological Data
Metagenomic Sequencing
Data Analysis
| Citation: |



 Download:
Download:




