-
The recent outbreak of novel coronavirus-infected pneumonia (NCIP) in China (1) has attracted much attention domestically and internationally. As the outbreak occurred, China carried out emergency responses quickly, notified the World Health Organization (WHO) in a timely manner, and shared the viral gene sequence with international communities immediately after pathogenic identification.
The timely release of information on the novel coronavirus (2019-nCoV), joint research on relevant viral genome mutations, pathogenicity, and transmission mechanisms, vaccine development, and drug screening and evaluation are of great significance for epidemic prevention and control. Therefore, for prevention and control of the NCIP epidemic and for future scientific research on 2019-nCoV, the National Pathogen Resource Center (NPRC) published information of the first strain of 2019-nCoV on the National Science and Technology Resource System for Novel Coronavirus (http://nmdc.cn/#/nCoV) on January 24, 2020.
HTML
-
Researchers from the National Institute for Viral Disease Control and Prevention (IVDC) of China CDC successfully isolated the virus out of lower respiratory alveolar lavage fluid from patients with unexplained viral pneumonia in Wuhan and detected typical electron microscopic coronavirus morphology on January 6, 2020, which substantiated the successful isolation and culture of the first 2019-nCoV strain in China (2).
-
Basic strain information of 2019-nCoV is important basis for downstream sharing, use, research, and epidemic prevention and control. With in-depth study of its biological traits and pathogenicity, further information will be supplemented gradually. (Table 1)
Descriptors Description Code CHPC 2020.00001, NPRC 2020.00001 Name in Chinese 新型冠状病毒武汉株01 Name in English C-Tan-nCoV Wuhan Strain Taxonomy Novel β genus coronavirus Source of the Specimen Clinical Patients Source of Collection Wuhan, Hubei Province, China Isolation Date Jan 6, 2020 Risk Level BSL-3 Contact Info ivdcolm@ivdc.chinacdc.cn, chpc@chinacdc.cn Note: CHPC refers to the Center for Human Pathogen Collection of China CDC, and NPRC refers to the National Pathogen Resource Center of China. Table 1. Basic information for the first strain of 2019-nCoV.
-
Item Sequence Target 1 (ORF1ab) Forward primer (F) CCCTGTGGGTTTTACACTTAA Reverse primer (R) ACGATTGTGCATCAGCTGA Fluorescent probes (P) 5’-FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1-3’ Target 2 (N) Forward primer (F) GGGGAACTTCTCCTGCTAGAAT Reverse primer (R) CAGACATTTTGCTCTCAAGCTG Fluorescent probes (P) 5’-FAM-TTGCTGCTGCTTGACAGATT-TAMRA-3’ Note: Results reading: negative (no Ct value or Ct=40); positive (Ct value <37, considered positive); and suspicious (if Ct value is between 37 and 40, repeat testing is recommended). If Ct value <40 in repeat testing and distinctive peaks are observed in the amplification curve, the sample is considered positive; otherwise, the sample is considered negative. Table 2. Real-time RT-PCR. The open reading frame 1ab (ORF1ab) and the nucleoprotein N gene region of the virus were recommended for primers and probes.
-
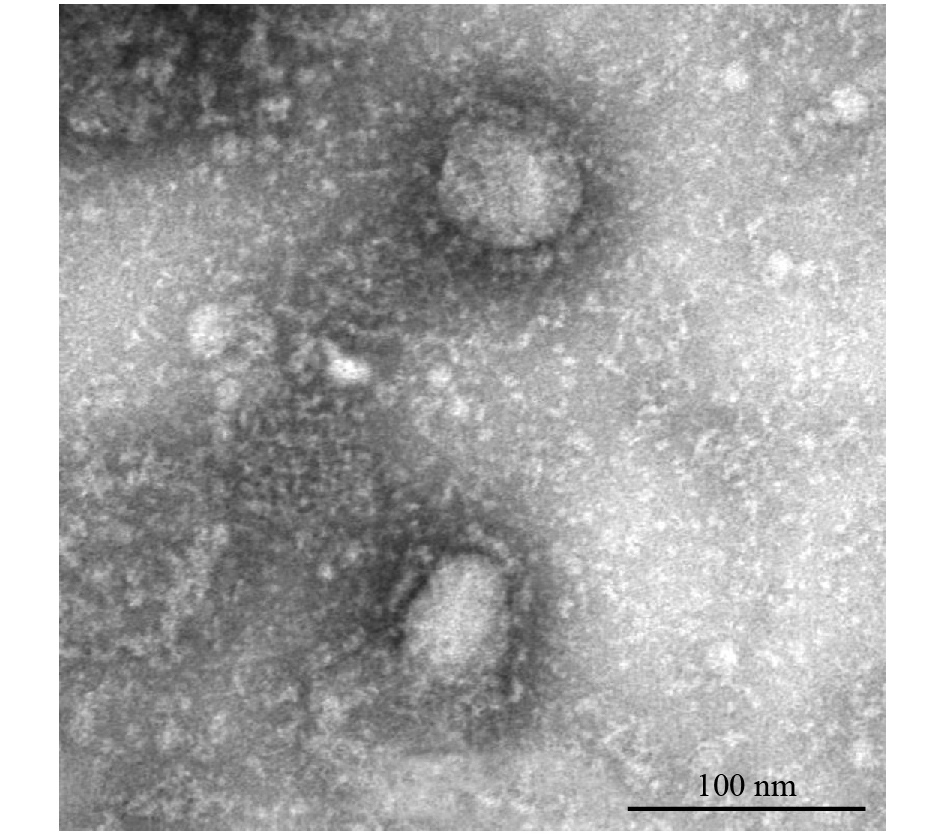
Negatively stained 2019-nCoV particles generally present a spherical shape with some pleomorphism. The viral particles range from 60–140 nm in diameter. Discernable spikes of 9–12 nm on the envelope render the virions appearance of a solar corona (hence named coronavirus) (Figure 1).
-
The 2019-nCoV strains can be shared for external use under proper administrative approvals and specified conditions. Users must indicate the source of the strains on relevant materials, including but not limited to publications and awards, as the following: National Pathogen Resource Center (National Institute for Viral Disease Control and Prevention, China CDC) with the NPRC number. Upon completion of the specified activities, users should return the strains to NPRC (IVDC) or perform proper destruction and disposal and issue a description of use to NPRC.
NPRC is a national science and technology resource sharing service platform established by China CDC. As a National Scientific and Technological Innovation Center, it is mainly responsible for the collection, identification, labeling, preservation, and provision of national pathogenic microbial resources. Through systemic integration of pathogenic microbial resources and enhancement of utility efficiency, NPRC is contributing to scientific and technological development as well as social welfare with its lasting conviction for a sharing and open society.
-
This work was supported by National Science and Technology Major Project on Important Infectious Diseases Prevention and Control (2018ZX10734404) and National Key Research and Development Project (2020YFC0840800, 2020YFC0840900).
| Citation: |



 Download:
Download:




