2023 Vol. 5, No. 26
The hospital-acquired infections caused by New Delhi metallo-beta-lactamase (NDM)-producing strains are typically attributed to a single clonal lineage.
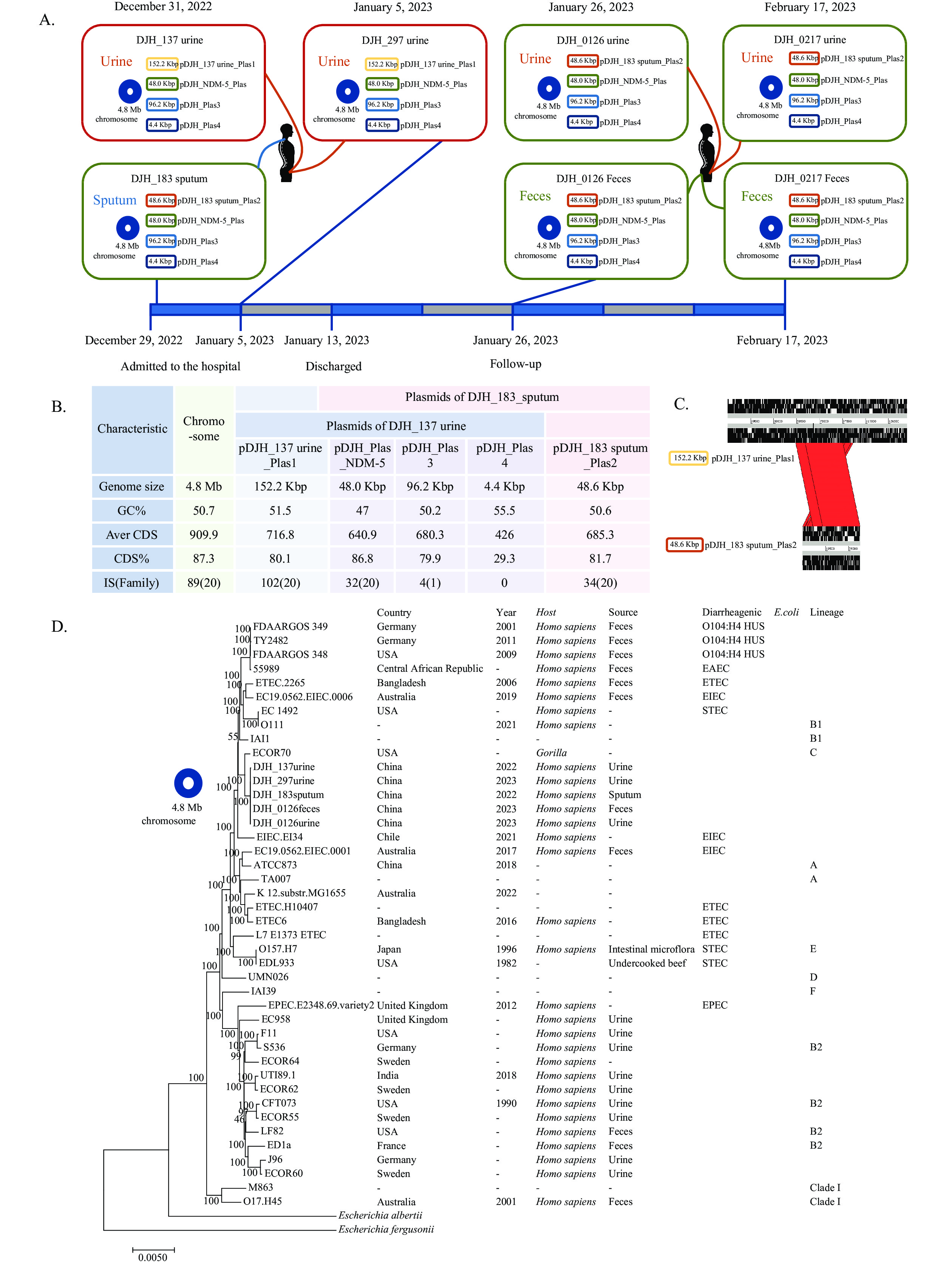
In this study, we encountered a unique case of community-acquired NDM-5 Escherichia coli urinary tract infection (UTI) following coronavirus disease 2019 (COVID-19). The UTI persisted for a duration of at least 45 days. Genomic analyses revealed the presence of two NDM-5 strains, both sharing an identical chromosomal background but distinct, homologous, and recombined plasmids. This case suggests that a diverse range of resistance genes may be present within the human body, with drug-resistant strains undergoing continuous evolution during infection. The intestinal tract may have been its drug-resistant gene pool.
The observations presented in this case indicate that the endogenous acquisition of drug-resistant genes may also be an issue in managing multidrug-resistant organisms (MDRO). It is possible for continuous recombination to occur within carbapenem-resistant Enterobacteriaceae (CRE) during infection. In contrast to exogenously-acquired resistance, greater attention should be placed on the endogenous factors that contribute to the development of CRE within healthcare settings.
In December 2022, China revised its epidemic prevention and control strategy, leading to an increase in coronavirus disease 2019 (COVID-19) cases and a peak in medical consultations. Government departments implemented relevant policies to coordinate and allocate medical resources throughout China. However, there is a scarcity of research on the status of medical consultations and the factors influencing them.
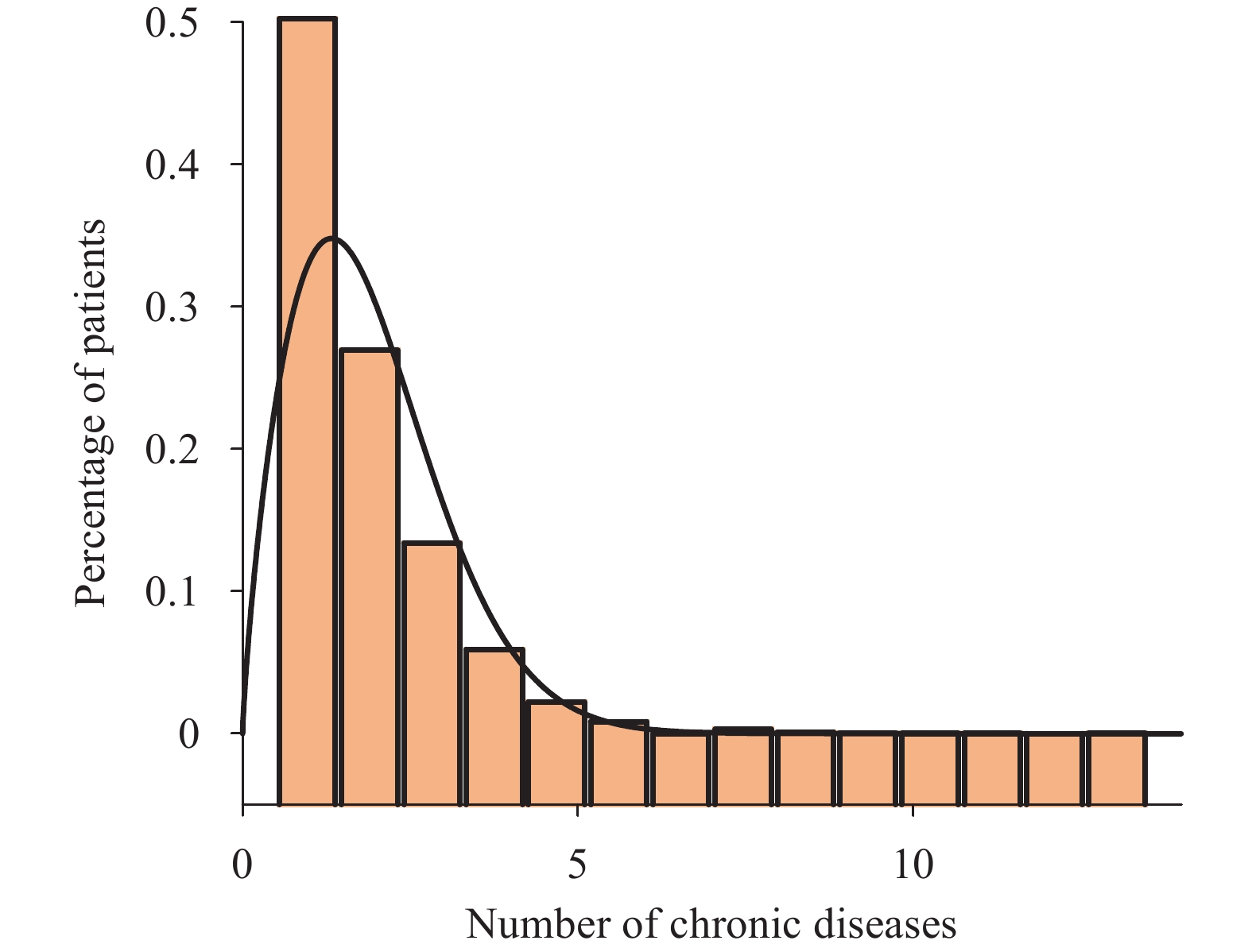
In the study population, over 80% of individuals with COVID-19 chose not to pursue medical care, while more than 70% of patients who sought treatment opted for primary healthcare facilities. The decision to consult medical professionals was influenced by various factors, such as age, education level, employment status, urban-rural distribution, and the presence of symptoms following COVID-19 infection.
The implementation of tiered diagnostic and treatment approaches, aligned with guidelines issued by governing bodies, is essential for mitigating the strain on medical resources. Primary healthcare institutions serve as “gatekeepers” for public health and should be further expanded in the future.
Since 2019, numerous variants of concern for severe acute respiratory syndrome virus 2 (SARS-CoV-2) have emerged, leading to significant outbreaks. The development of novel, highly accurate, and rapid detection techniques for these new SARS-CoV-2 variants remains a primary focus in the ongoing efforts to control and prevent the coronavirus disease 2019 (COVID-19) pandemic.
Reverse transcription-recombinase polymerase amplification combined with the clustered regularly interspaced short palindromic repeats-associated protein 12a (CRISPR/Cas12a) system was used to validate the detection of the Omicron BA.2, BA.4, and BA.5 variants of SARS-CoV-2.
Our results demonstrate that the CRISPR/Cas12a assay is capable of effectively detecting the SARS-CoV-2 BA.2, BA.4, and BA.5 variants with a limit of detection of 10, 1, and 10 copies/μL, respectively. Importantly, our assay successfully differentiated the three SARS-CoV-2 Omicron strains from one another. Additionally, we evaluated 46 SARS-CoV-2 positive clinical samples consisting of BA.2 (n=20), BA.4 (n=6), and BA.5 (n=20) variants, and the sensitivity of our assay ranged from 90% to 100%, while the specificity was 100%.
This research presents a swift and reliable CRISPR-based method that may be employed to track the emergence of novel SARS-CoV-2 variants.



 Subscribe for E-mail Alerts
Subscribe for E-mail Alerts CCDC Weekly RSS Feed
CCDC Weekly RSS Feed