-
Ongoing surveillance of severe acute respiratory syndrome virus 2 (SARS-CoV-2)’s genetic variations remains crucial for effective epidemic prevention and control. Our prior research demonstrates the co-circulation of two pre-existing Omicron subvariants, BA.5.2 and BF.7, which were responsible for the coronavirus disease 2019 (COVID-19) surge in Beijing following modifications to prevention and control policies in mid-November 2022 — a snapshot of China’s situation at the time (1). Following a period of relatively low COVID-19 case count after February 2023, a noticeable increase was observed from late April 2023 (2). Post-analysis of SARS-CoV-2 genomic data gathered in China post November 11, 2022, [sourced from Global Initiative of Sharing All Influenza Data (GISAID) as of July 6, 2023], revealed that the Omicron subvariants most detected were BA.5.2, BF.7, DY (descendants of BA.5.2.48), and XBB — along with their corresponding descendants denoted as BA.5.2*, BF.7*, DY*, and XBB*, respectively (Figure 1A). Interestingly, detection of BA.5.2*, BF.7*, and DY* plateaued by May 2023, while that of XBB*, previously without local transmission, showed notable growth post-April 2023 (Figure 1B). Further, key nonsynonymous mutations were identified, with high-frequency mutations (≥10%) being 4 in BA.5.2*, 4 in BF.7*, 6 in DY*, and 7 in XBB*, excluding the defining mutations present in ≥80% of the genomes in each lineage (Figure 1C). Six of these mutations showed high prevalence in China (frequency ≥80%). However, these 21 mutations were not exclusive to China, having been first identified outside the country prior to 2023 (Figure 1D). Collectively, despite the co-circulation of BA.5.2*, BF.7*, and DY* in China for over six months, prevalently novel nonsynonymous mutations have not been discovered in them. Notably, XBB* has displayed significant immune-evasion capabilities (3) and has effectively evaded responses induced by several types of vaccines widely used in China (4). Moreover, it has been approximately six months since the first large-scale infections wave at the end of 2022, with patient antibody levels gradually declining, implying the dominance of XBB* over BA.5.2*, BF.7*, and DY* and triggering a second wave of infections. Given China’s dense population, ongoing local transmissions — both local and imported — of various SARS-CoV-2 variants have the potential to generate innovative variants in humans or animals. Consequently, continuous, comprehensive surveillance remains vital and should be conducted under the “One Health” framework.
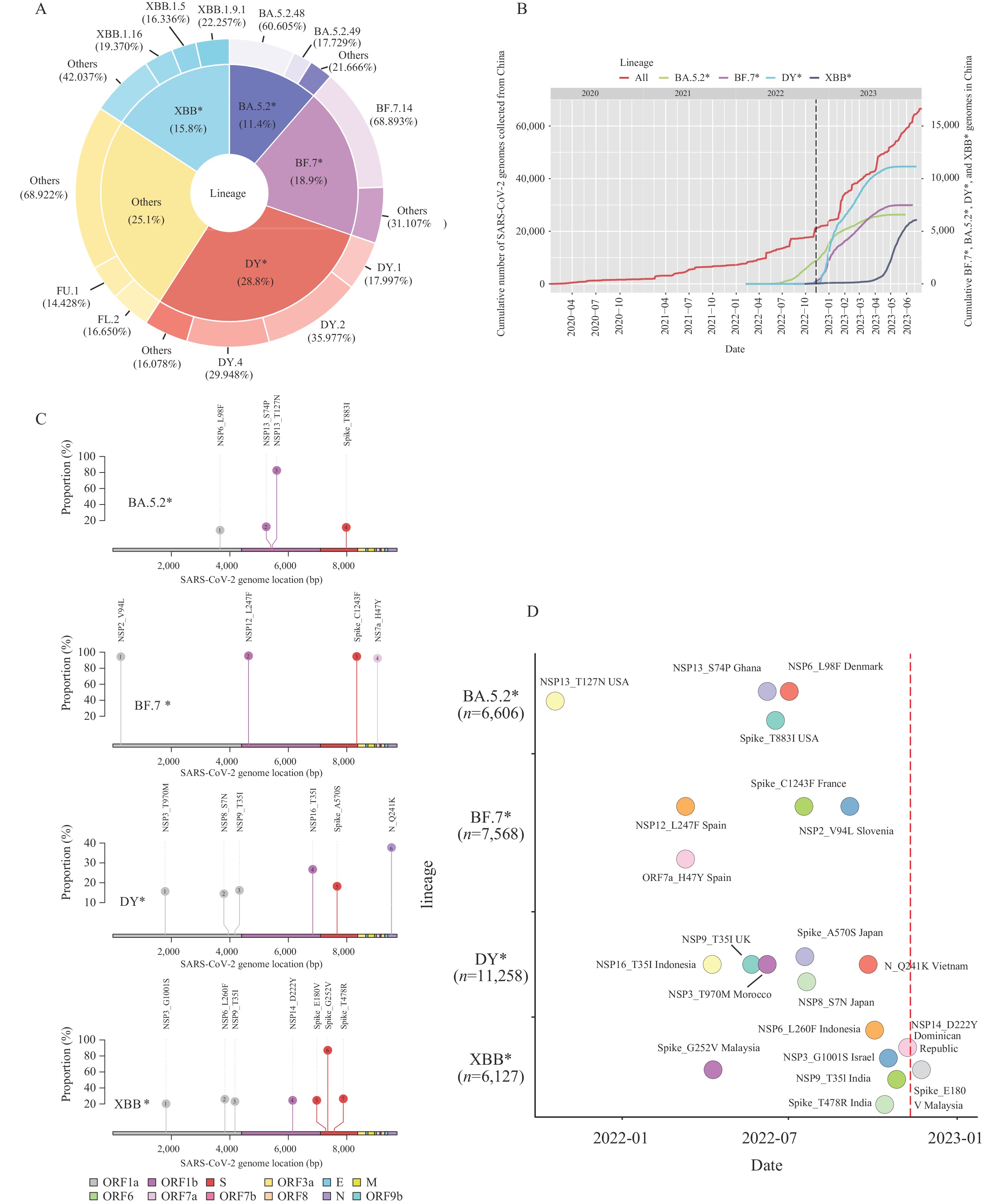 Figure 1.
Figure 1.Trends and statistics of BA.5.2*, BF.7*, DY*, and XBB* variants of severe acute respiratory syndrome virus 2 (SARS-CoV-2) genomes in China. (A) The composition of SARS-CoV-2 variants circulating from November 11, 2022, to June 30, 2023. (B) The cumulative number of SARS-CoV-2 genomes derived from China. (C) Mutations categorized as BA.5.2*, BF.7*, DY*, and XBB* have been identified with high frequency in China. (D) The initial detection date and location of each high-frequency mutation within their respective lineages.
Note: Different lineages are demonstrated through color-coding. The red dashed line represents the timing of preventive and control policy adjustments. The number of genomes gathered from China for each lineage is also depicted. The implementation of changes to prevention and control policies is represented by a dashed line in panel B. Genes encoded by SARS-CoV-2 genomes were demonstrated through color-coding in panel C. -
George F. Gao is the Founding Editor-in-Chief of China CDC Weekly. He was not involved in the peer review or handling of the manuscript. The authors have no other competing interests to disclose.
-
We gratefully acknowledge the data contributors where SARS-CoV-2 genomic sequence data were generated and shared via GISAID.
HTML
| Citation: |




 Download:
Download:




