-
On July 4, 2022, the first case of Omicron subvariant BA.5.2 in Beijing Municipality was discovered in Yanqing District. The case was a 49-year-old Chinese male who had arrived in Shanghai Municipality via international flight DL9927 from North Carolina, U.S. on June 15. He stayed in a hotel for the 14-day arrival quarantine and was discharged on June 30. He arrived in Beijing via domestic flight MU5103 on July 1 and was transferred point-to-point from the airport to his residence community in Yanqing District. On July 3, his sample was collected through community mass screening and reported positive in the next morning. The case had received 3 doses of Moderna’s mRNA vaccines in the U.S., with the last shot on May 26, 2022.
The index case caused an outbreak with a total of 16 cases located in 5 districts within 7 days. Among all cases, 12 were males and 4 were females; 12 received a booster shot, whereas 4 were non-vaccinated.
The respiratory samples from 16 cases were sequenced by the Next Generating Sequencing (NGS). A total of 13 full genomes were obtained and all belonged to the same lineage BA.5.2, Variant of Concern (VOC)/Omicron. In particular, 11 genomes shared a nucleotide similarity of 100%, 1 genome carried an additional mutation of T29678C, and 1 carried an additional heterozygous mutation of A6821G with a frequency of 59.39%. Phylogenetic analysis indicated the virus was similar to strains in North America, Europe, and Asia in mid-June, which was different from local clusters in Beijing in the same period (Figure 1).
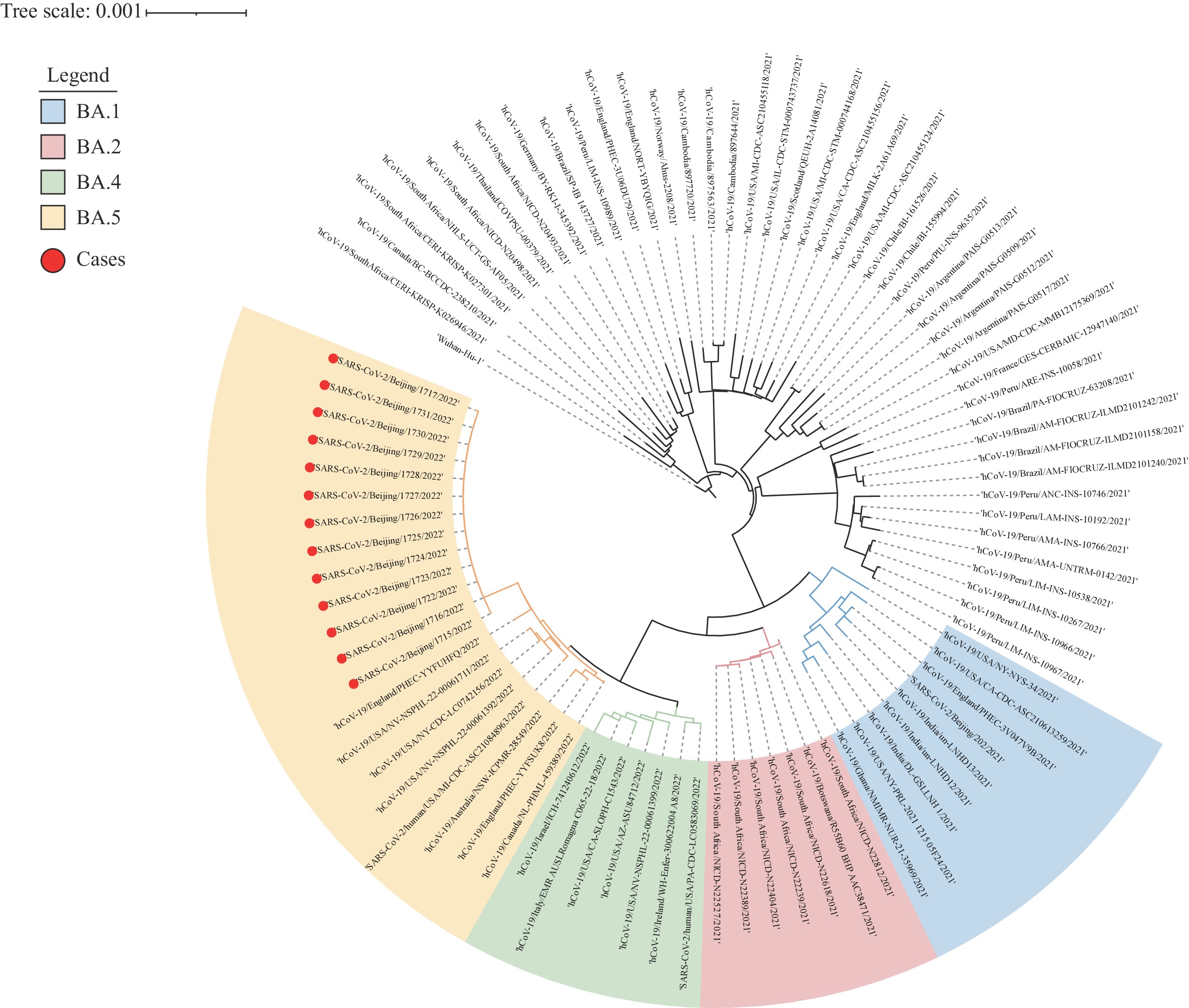 Figure 1.
Figure 1.Neighbor-joining phylogenetic tree of strains in the outbreak.
Note: The genomes from cases in the outbreak were indicated by red dots. The major VOC/Omicron PANGOLIN lineages were marked and colored on the right. The tree was rooted using strain Wuhan-1 (EPI_ISL_402125).
Abbreviation: VOC=variant of concern.
Omicron subvariant BA.5 had surged dramatically to become dominant in the U.S. (1). Research indicated that the Omicron BA.5 subvariant had a growth advantage against other subvariants with a higher ability of immune escape than the BA.1, BA.2, and BA.2.12.1 subvariants (2-5). Continuous surveillance and assessment need to be implemented to respond Omicron subvariant BA.5 in China.
HTML
| Citation: |




 Download:
Download:




