-
On December 9, 2021, 2 international passengers arrived in Tianjin Binhai International Airport from Warsaw, Poland via airplane and tested positive for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease 2019 (COVID-19), using nucleic acid tests. The first case (Patient A) was a flight crew member, a 35-year-old male, who had returned to Warsaw on the same flight on December 10, 2021. The second case (Patient B) was a 17-year-old female student, who has been transferred to Haihe Hospital for isolation. Both cases are Polish nationality that had recently lived in Warsaw, Poland. According to the investigation, neither patient had disease symptoms nor medication history. Patient B received the Pfizer/BNT162b2 vaccine on September 27, 2021. Patient A was vaccinated on May 25, 2021, but no further vaccine information was collected as Patient A had left China. No other infections from this flight have been detected.
On December 10, 2021, Tianjin CDC received specimens from the 2 cases, sequenced using Illumina iSeq platform (Illumina, San Diego, CA, USA), and obtained the sequencing results on December 13. Compared with the Wuhan reference (EPI_ISL_402125) (1-2), the viral nucleotide sequence from Patient A displayed 58 substitutions, 39 deletions, and 9 insertions (genomic coverage 99.62%) and belonged to Pango lineage BA.1 (alias of B.1.1.529.1, GISAID ID: EPI_ISL_7734647) (Figure 1). Due to low viral load, the coverage of the viral genome from Patient B was only 92.62%. Based on the characteristics of the detected 50 substitutions and 3 deletions, the strain from Patient B belonged to lineage B.1.1.529. The lineage B.1.1.529 and its descendants (BA.1 and BA.2) were designated as the fifth SARS-CoV-2 Variant of Concern (VOC) by World Health Organization (WHO), named Omicron, following the designation of the Alpha, Beta, Gamma, and Delta variants. This variant was first reported to the WHO by South Africa on November 24, 2021. As of December 14, 2021, 55 counties shared 4,265 Omicron genome sequences in the GISAID database (3).
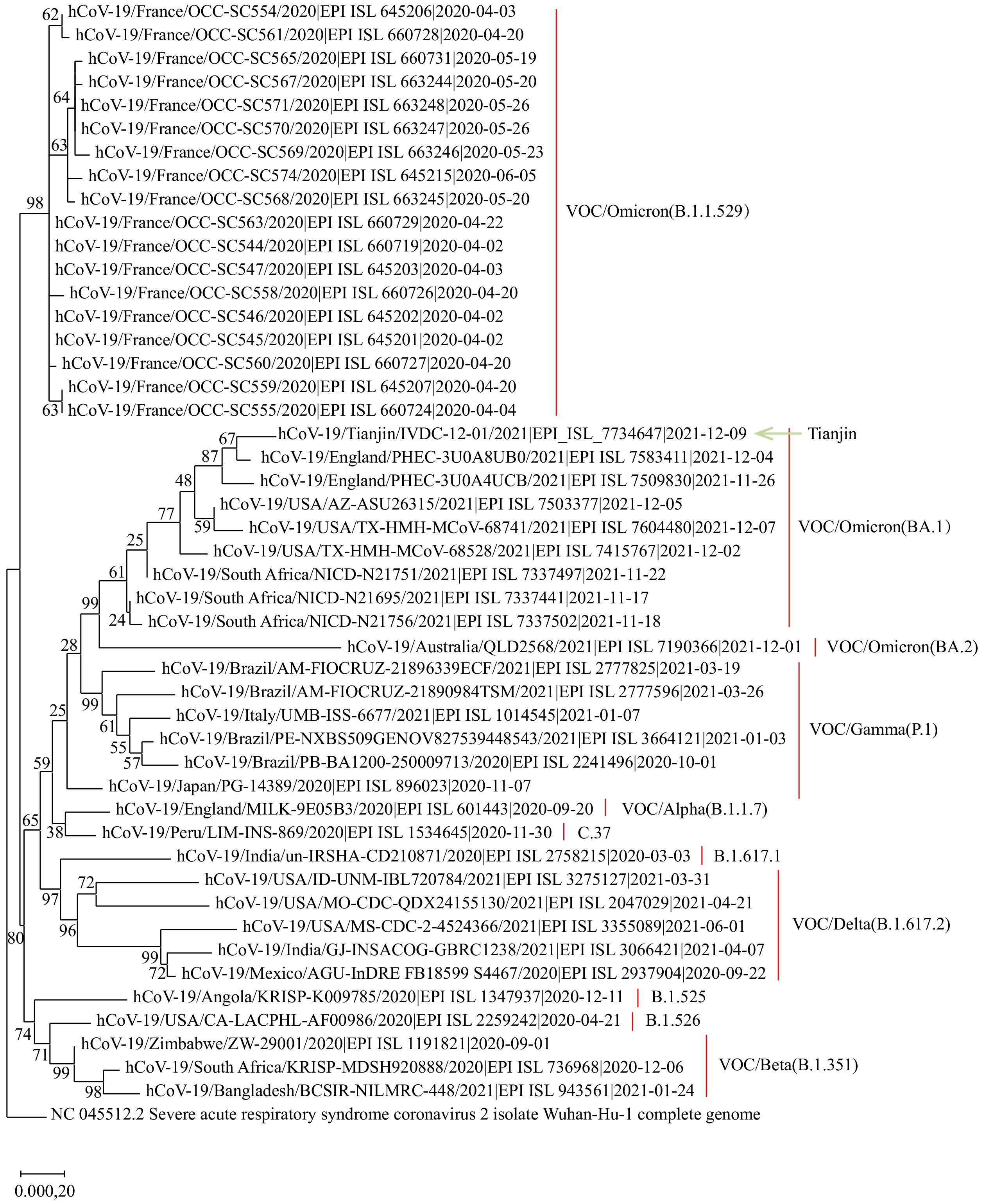 Figure 1.
Figure 1.Phylogenetic tree based on the full-length genome sequences of SARS-CoV-2.
Notes: The Tianjin imported Omicron variant is marked with a green arrow. The three lineages of the Omicron variant (GISAID ID: EPI_ISL_7734647) and other seven distinguished SARS-CoV-2 variants are marked and colored on the right. The Wuhan reference strain is on the bottom. The nucleotide sequence from Patient B is not included as its genomic sequence is incomplete.Abbreviations: SARS-CoV-2=severe acute respiratory syndrome coronavirus 2; VOC=variant of concern.A total of 40 amino acid mutation sites (A67V, T95I, Y145D, L212I, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, A701V, N764K, D796Y, N856K, Q954H, N969K, L981F, H69del, V70del, G142del, V143del, Y144del, N211del, and 214insEPE) and at least 26 amino acid mutation sites (T19R, G142D, C361R, K424I, N439I, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, F497Y, Q498R, N501Y, Y505H, N679K, P681H, A701V, N764K, D796Y, N856K, Q954H, N969K, L981F, and P1069S) were detected in the spike protein of the strains from Patient A and B, respectively. Some of the mutations in the receptor binding domain and near the furin cleavage were concerning and may be associated with immune escape potency and higher transmissibility (4).
Nowadays, Omicron is displaying a growth advantage over other circulating variants in many countries of the world (5-6). In addition, preliminary laboratory data have shown that Omicron displayed a reduction of immune protection against infection and vaccine (7). The strains from two cases in Tianjin were the first detected cases of the imported Omicron variant in the mainland of China and pose a great potential threat to the prevention and control of COVID-19 in China. The transmissibility, pathogenicity, and immune evasion of Omicron urgently needs to be further studied.
HTML
| Citation: |



 Download:
Download:




