-
Tales of tracing the origins of human immunodeficiency virus (HIV), human coronavirus HKU1 (HCoV-HKU1), severe fever with thrombocytopenia syndrome virus (SFTSV), and Middle East respiratory syndrome coronavirus (MERS-CoV) can enlighten the scientific search for the origins of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Here, we detail key research studies on the origins of these four viruses.
On June 5, 1981, US CDC reported five cases of now-called acquired immunodeficiency syndrome (AIDS) in the Morbidity and Mortality Weekly Report (MMWR) — the first official report of AIDS in the world (1). Two years later, scientists at the Pasteur Institute in France isolated the pathogen, HIV, from lymphoid ganglions (2). Last year was the 40th anniversary of the initial report of AIDS, but we still lack a successful vaccine for its prevention. In 1986, Nahmias and colleagues conducted serological studies of 1,213 plasma samples that were obtained as early as 1959 from various parts of Africa, and confirmed with indirect immunofluorescence assay (IFA), Western blot, and radioimmunoprecipitation that one of the samples was positive for HIV-1 (3). The positive sample had been collected in early 1959 from an adult Bantu member who had glucose-6-phosphate dehydrogenase deficiency and was living in Léopoldville, now Kinshasa, Democratic Republic of Congo (4). In 1998, Zhu and colleagues reported the HIV-1 nucleotide sequence from this 1959 sample. Phylogenetic analysis confirmed HIV-1 infection, and the phylogenetic position of the virus was near the ancestor node of HIV-1 subtypes B, D, and F, suggesting that these HIV-1 subtypes may have evolved from an introduction into the African population prior to 1959 (5). A 2008 report further pushed back the putative origins of HIV by several decades. Worobey and colleagues performed viral genome sequencing on a Bouin’s-fixed, paraffin-embedded lymph node biopsy specimen obtained in 1960 from an adult female in Léopoldville. Evolutionary analysis of the virus sequence in the specimen dates the most recent common ancestor (TMRCA) of HIV-1 M group to 1908 (95% confidence interval: 1884–1924) — about a hundred years before recognition of the AIDS epidemic (6) (Figure 1A).
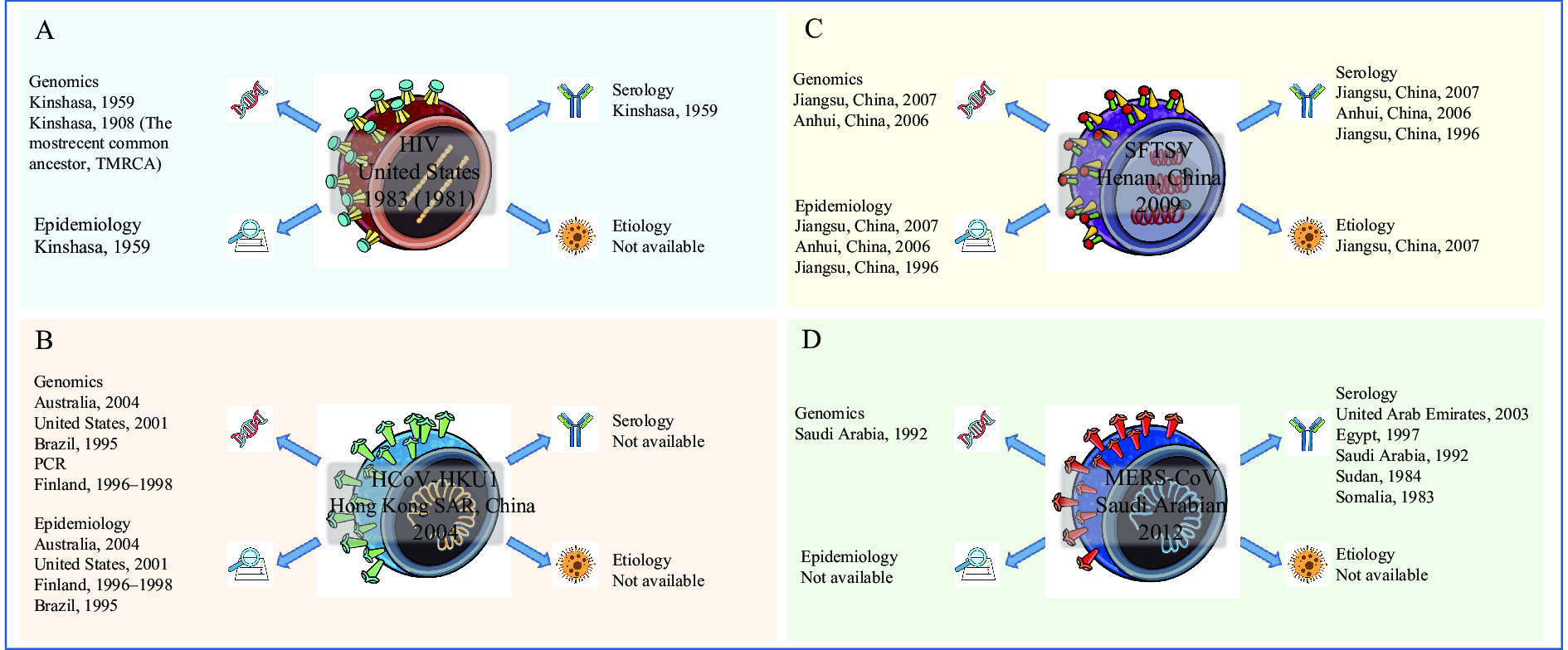 Figure 1.
Figure 1.The origins-tracing of (A) HIV, (B) HCoV-HKU1, SFTSV, and MERS-CoV.
Note: Graphs illustrate the time and place of the first discovery of HIV, HCoV-HKU1, SFTSV and MERS-CoV, as well as the tracing history of these viruses from four aspects: genomics (i.e., with nucleic acid sequencing or specific PCR products), epidemiology, serology, and etiology (virus isolation). (A) HIV was first reported in the United States in 1981 and the virus was isolated in 1983; it has been traced back to Kinshasa in 1908. Except for etiology, all genomics, serology, and epidemiology could be traced back to 1959. (B) HCoV-HKU1 was first identified in Hong Kong, China in 2004 and has been traced back to Brazil in 1995. Other evidence of genomics and epidemiology was observed in other countries before 2004, as shown. (C) SFTSV was first discovered in Henan Province, China in 2009 and has been traced back to Jiangsu Province, China, as early as in 1996. Some additional evidence in 2006 and 2007 was seen. (D) MERS-CoV was first reported in Saudi Arabia in 2012 and has been traced back to Somalia as early as 1983. Other evidence between 1983 and 2012 is shown in the figure.
Abbreviation: HIV=human immunodeficiency virus; HCoV-HKU1=human coronavirus HKU1; SFTSV=severe fever with thrombocytopenia syndrome virus; MERS-CoV=Middle East respiratory syndrome coronavirus; PCR=polymerase chain reaction.
Coronavirus HCoV-HKU1, which causes human respiratory tract infections, was first identified in 2004 in a 71-year-old male returning to Hong Kong SAR, China from Shenzhen City, Guangdong Province. The virus was named after the University of Hong Kong, where it had been discovered (7). In 2006, researchers detected HCoV-HKU1 positive polymerase chain reaction (PCR) signals in specimens from Australian children suffering from upper or lower respiratory tract illnesses in autumn or winter of 2004 (8). Also in 2006, using reverse transcription-PCR (RT-PCR), American researchers identified HCoV-HKU1 in children’s respiratory specimens collected in Connecticut, USA during 2001 and 2002 — the first identification of HCoV-HKU1 in the Western Hemisphere (9). In 2009, Finnish researchers discovered HCoV-HKU1 using RT-PCR in nasopharyngeal aspirates collected from Finnish children between 1996 and 1998 (10). In a retrospective study in Brazil, scientists used a universal coronavirus PCR assay and identified HCoV-HKU1 in children’s nasopharyngeal swab samples that were frozen and stored in 1995, thus pushing back HCoV-HKU1 identification to 1995 (11). Together, these results show that HCoV-HKU1 was present in Europe and the Americas before its discovery in 2004 in Hong Kong SAR, China (Figure 1B).
In 2009, scientists in Henan Province, China made the initial discovery of SFTSV in patients with severe fever with thrombocytopenia syndrome (SFTS) (12). This discovery prompted researchers in Jiangsu Province to perform SFTSV testing on samples obtained in 2007 from patients with similar clinical manifestations and elusive etiologies (13). Six blood specimens tested positive for SFTSV RNA by real-time RT-PCR and positive for SFTSV antibody by microneutralization assay (MNA) and IFA. In addition, SFTSV was isolated from one serum specimen. In 2012, researchers performed SFTSV IFA and RT-PCR tests on sera collected in 2006 from 13 patients suffering from infections of unknown etiology in Anhui Province, China. Tests for SFTSV were positive, and given that all secondary patients had contact with blood from the index patient or underwent endotracheal intubation, the test results suggested that the virus could spread from person to person through contact with blood (14). Another group of researchers performed SFTSV testing on sera obtained in 1996 from six patients with SFTS in Yixing County, Jiangsu Province (15). Enzyme-linked immunosorbent assays (ELISA) identified IgM antibodies against SFTSV in sera from all six patients, while IFA testing found IgG antibodies in one patient. This study demonstrated that SFTSV IgG antibodies could still be detected in the sera 14 years after disease onset. Based on epidemiological analyses, clinical symptoms, and serological studies, unexplained fever and thrombocytopenia were determined to have been caused by SFTSV. Taken together, these studies demonstrated that SFTSV was in China more than ten years before it was first discovered in 2009 (Figure 1C). Based on phylogenetic analyses of SFTSV genomics, the scientists concluded that SFTSV originated in the early 18th century from Zhejiang Province and that Genotype F was an early genotype, thus promoting a more comprehensive understanding of the origin of SFTSV (16). This experience also shows that the discovery of the emergence of a new pathogen in the human population and the origin tracing of a new pathogen can be separate scientific events.
MERS-CoV has spread from the Arabian Peninsula to over 20 countries in Europe, Africa, Asia, and North America since it was first reported in a 60-year-old Saudi Arabian man who died in 2012; these cases were all sporadic, unable to be linked into chains of transmission (17). Azhar and colleagues suggested that MERS-CoV can be transmitted from camels to humans through close contact, and that camels may act as intermediate hosts that transmit the virus from its reservoir to humans (18). Bats are proposed to be a reservoir host, as partial genomic sequences with 100% identity to MERS-CoV were discovered in bat samples dating back to 2012 (19). A study by Meyer and colleagues published in 2014 analyzed 651 dromedary camel serum samples from the United Arab Emirates using recombinant spike protein-specific IFA and virus neutralization tests. A total of 151 samples had been collected in 2003 (20). The study found that 97.1% of dromedaries (632 of 651 samples, including all of the dromedary sera collected in 2003) had antibodies against MERS-CoV, and that 59.8% of serum samples had high MERS-CoV neutralizing antibody titers — greater than 1,280. Antibodies discovered in the serum samples that were obtained in 2003 indicated that a high proportion of dromedaries in the region were infected with MERS-CoV or a conspecific virus long before the first human case was identified. In 2013, Lipkin and colleagues performed ELISA, Western blot, luciferase immunoprecipitation system (LIPS) assays, and nucleotide sequencing on freshly collected dromedary camels, sheep, and goat samples and on archived serum samples collected during 1992–2010 in the Kingdom of Saudi Arabia. Their results suggested that MERS-CoV had been circulating countrywide in camels for at least 2 decades, and that it had evolved into phylogenetic clades related to human infections (21). In 2014, Müller and colleagues used a highly specific MERS-CoV microneutralization assay to test 189 archived dromedary serum samples accumulated over the previous 30 years, including sera collected in Somalia during 1983–1984, in Sudan June–July 1984, and in Egypt June–July 1997. They found that 81.0% of samples were positive for MERS-CoV antibodies, indicating that the virus had been circulating in these animals for decades before its discovery in 2012 (22) (Figure 1D).
Similarly, multiple lines of evidence show that before the coronavirus disease 2019 (COVID-19) outbreak in Wuhan Huanan Seafood Wholesale Market, sporadic positive samples and cases of SARS-CoV-2 appeared in many countries and regions. In October 2021, researchers from IRCCS National Cancer Institute Foundation reported multiple SARS-CoV-2 antibody positive serum samples collected in Milan, Italy starting from September 2019 (23). In December 2020, researchers from the University of Milan reported a positive test for SARS-CoV-2 in an oropharyngeal swab sample collected on December 5, 2019 from a 4-year-old boy with no prior travel history (24). In January 2021, another group of researchers from the University of Milan reported that the SARS-CoV-2 gene sequence was detected in a biopsy sample collected from a 25-year-old female patient with skin disease in Italy on November 10, 2019 (25). In a preprint of The Lancet released on August 6, 2021, Amendola and colleagues collected 435 oropharyngeal swabs and urine and serum samples from 156 individuals with morbilliform rashes and tested them for SARS-CoV-2 infection by PCR, Sanger sequencing, ELISA, and SARS-CoV-2 plaque reduction neutralization assays. The first positive result of SARS-CoV-2 RNA was found in a sample that was collected in September 2019. Researchers estimated that SARS-CoV-2 progenitors emerged in late June to late August 2019 (26). These results confirmed that the virus had been prevalent in Italy before the official announcement of the first confirmed local COVID-19 case on February 21, 2020.
In April 2020, the mayor of Belleville, New Jersey, Michael Melham, announced that he had tested positive for antibodies against SARS-CoV-2 and believed that he had been infected with the virus in November 2019 (27), even though the first confirmed case of COVID-19 in the United States was identified on January 21, 2020. In November 2020, US CDC researchers reported that they tested 7,389 blood samples collected by the American Red Cross between December 13, 2019 and January 17, 2020 and found 106 blood samples containing antibodies against SARS-CoV-2 (28). In June 2021, US National Institutes of Health (NIH) (29) and ABC NEWS (30) both reported a study initiated by NIH in which scientists tested 24,000 blood samples that were collected in early 2020 across the United States. The study found that SARS-CoV-2 antibodies were detected in blood samples from at least 9 people, with the earliest positive sample collected on January 7, 2020. Since antibodies do not appear until about two weeks after human infection, this finding suggests that SARS-CoV-2 was circulating at a low level in the United States as early as December 2019 (31).
Researchers from Paris Seine Saint-Denis Hospital Group, Bobigny, France retrospectively tested a respiratory specimen obtained from a patient with hemoptysis in December 2019 and confirmed that the patient was infected by SARS-CoV-2. Judging from this result, the outbreak in France started earlier than the official notification of the first confirmed case on January 24, 2020 (32).
Worldwide, earlier SARS-CoV-2 infections have not only been found in human cases, but also, studies of wastewater have shown that SARS-CoV-2 infections may have existed much earlier than the first reported human cases. The Italian Istituto Superiore di Sanità announced that SARS-CoV-2 was identified in wastewater samples collected in northern Italian cities Milan and Turin on December 18, 2019 — more than two months before the first local case of COVID-19 in the country (33). The Federal University of Santa Catarina in Brazil published a paper in which researchers detected SARS-CoV-2 RNA in human sewage samples collected in Florianópolis, Brazil on November 27, 2019, about three months before the first case of COVID-19 was reported in Brazil (34). Similarly, an investigation in Spain found that SARS-CoV-2 was detected in a frozen wastewater sample collected from a wastewater treatment plant in Barcelona on January 15, 2020, which was 41 days before the first confirmed case was reported in Spain (35).
Tracing the origin of a virus is scientific work, and solid conclusions only result from an enormous amount of effort, patience, global cooperation, some luck, and possibly decades of continuous research (36). There are common phenomena seen during the process of back-tracing a virus, including that transmission of a new pathogen is often greater in dense populations than in sparse populations. Therefore, emergence of major, new infectious diseases correlate strongly with human population density. Disease emergence is driven by the diagnostic ability of local doctors, the research capacity of scientists, the surveillance capabilities of local government, and the willingness to share information.
HIV was first reported in the United States in 1981 and has been traced back to Kinshasa in 1908. HCoV-HKU1 was first identified in Hong Kong SAR, China in 2004 and has been traced back to Brazil in 1995. SFTSV was first discovered in Henan Province, China in 2009 and has been traced back to Jiangsu Province, China in 1996. MERS-CoV was first reported in Saudi Arabia in 2012 and has been traced back to Somalia in 1983. Now, there is a growing number of clues, reports, and studies indicating that COVID-19 outbreaks occurred in multiple locations around the world before 2020. We should be inspired by the origin studies of previous viruses and carry out global cooperation to test more samples from patients that had COVID-19 symptoms, more environmental samples, and more susceptible-animal samples on larger spans of time and space.
HTML
| Citation: |




 Download:
Download:




