-
On April 23, 2022, an international flight KQ880 from Nairobi, Kenya arrived at Guangzhou Baiyun International Airport, Guangzhou City, Guangdong Province. All passengers were transferred to the quarantine hotel for routine 14-day medical observation and regularly tested of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleic acid. One of the patients, a 27-year-old Chinese male, reported positive SARS-CoV-2 nucleic acid on April 27, 2022. The patient had a history of full coronavirus disease 2019 (COVID-19) vaccination and denied exposure to other COVID-19 cases in the past 14 days. After diagnosis, he was transferred to Guangzhou Eighth People’s Hospital for treatment.
On April 30, 2022, a nasal swab sample from the patient was sequenced using the Illumina MiniSeq platform (Illumina, San Diego, CA, USA), and genotyping results showed that the patient was infected with Omicron subvariant BA.2.12.1. A total of 33 amino acid mutation sites (T19I, A27S, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452Q, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, S704L, N764K, D796Y, Q954H, N969K and L24del, P25del, P26del) were detected on the spike gene, two of which (L452Q and S704L) were key sites defining the signature of sublineage BA.2.12.1 (1-2) (Figure 1). The sequence has been submitted to the National Genomics Data Center (under the accession number WGS025539).
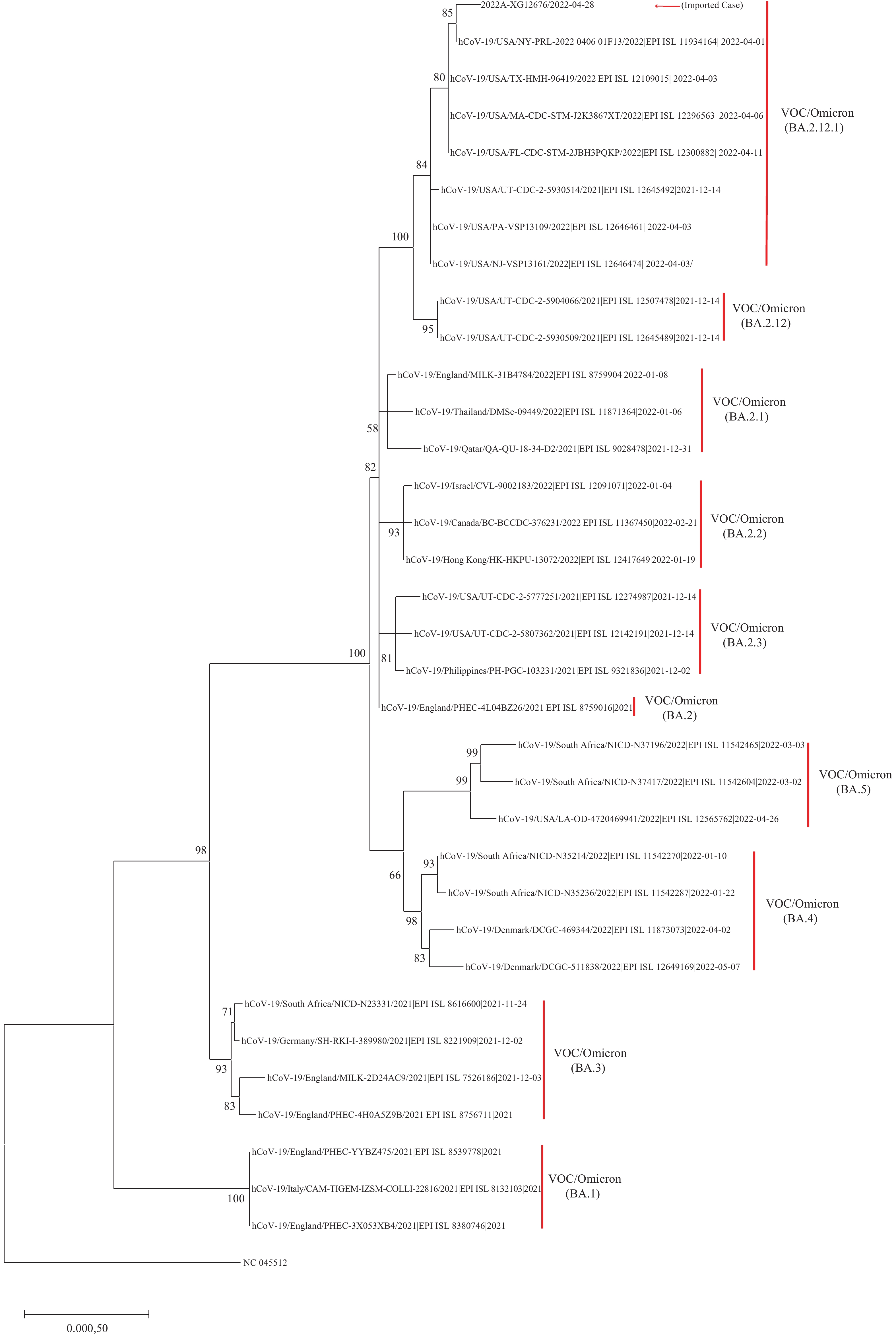 Figure 1.
Figure 1.Phylogenetic tree based on the full-length genome sequences of the COVID-19 Omicron subvariants.
Note: Guangdong Province imported VOC/Omicron (BA.2.12.1) variant is indicated with red arrow. Other Omicron sublineages are indicated on the right side and marked with square brackets.
Abbreviation: COVID-19=coronavirus disease 2019; VOC=variant of concern.
On May 4, 2022, the World Health Organization reminded to closely monitor BA.2.12.1 subvariant (2). Compared to other Omicron variants, BA.2.12.1 subvariant shows stronger immune escape, even after having been vaccinated with booster dose (3). Several studies have shown that the transmissibility of BA.2.12.1 is about 23% to 27% faster than that of BA.2. Omicron BA 2.12.1 subvariant spread very fast, which led to the resurgence of the epidemic in many parts of the United States, and cases have been reported in at least 17 countries (4). Public health officials are focusing on this subvariant and trying to learn more.
HTML
| Citation: |




 Download:
Download:




