-
Superspreaders were defined as the minority of individuals who infect disproportionately more susceptible contacts, especially when compared to the majority of individuals who infect few or no others. Superspreading events, i.e. events involving superspreaders, highlight the limitations of basic reproduction number (R0), which represented the average dynamics of transmission. Previous reports of superspreading events of COVID-19 indicate that these events contributed significantly to the rapid transmission of infections (1). During recent severe outbreaks of the severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and Ebola virus disease, superspreading events were associated with explosive growth early in an outbreak and sustained transmission in later stages (2). To determine how the virus may have spread in a superspreading event of COVID-19 associated with a hospital in Qingdao City, Shandong Province, China, we monitored and traced close contacts and hypothesized possible transmission modes. Real-time reverse transcription polymerase chain reaction (real-time RT-PCR) diagnosis based on nasopharyngeal swab was used for confirmation of this disease (3). The study was approved by Qingdao CDC, and all patients in this study were anonymized.
HTML
-
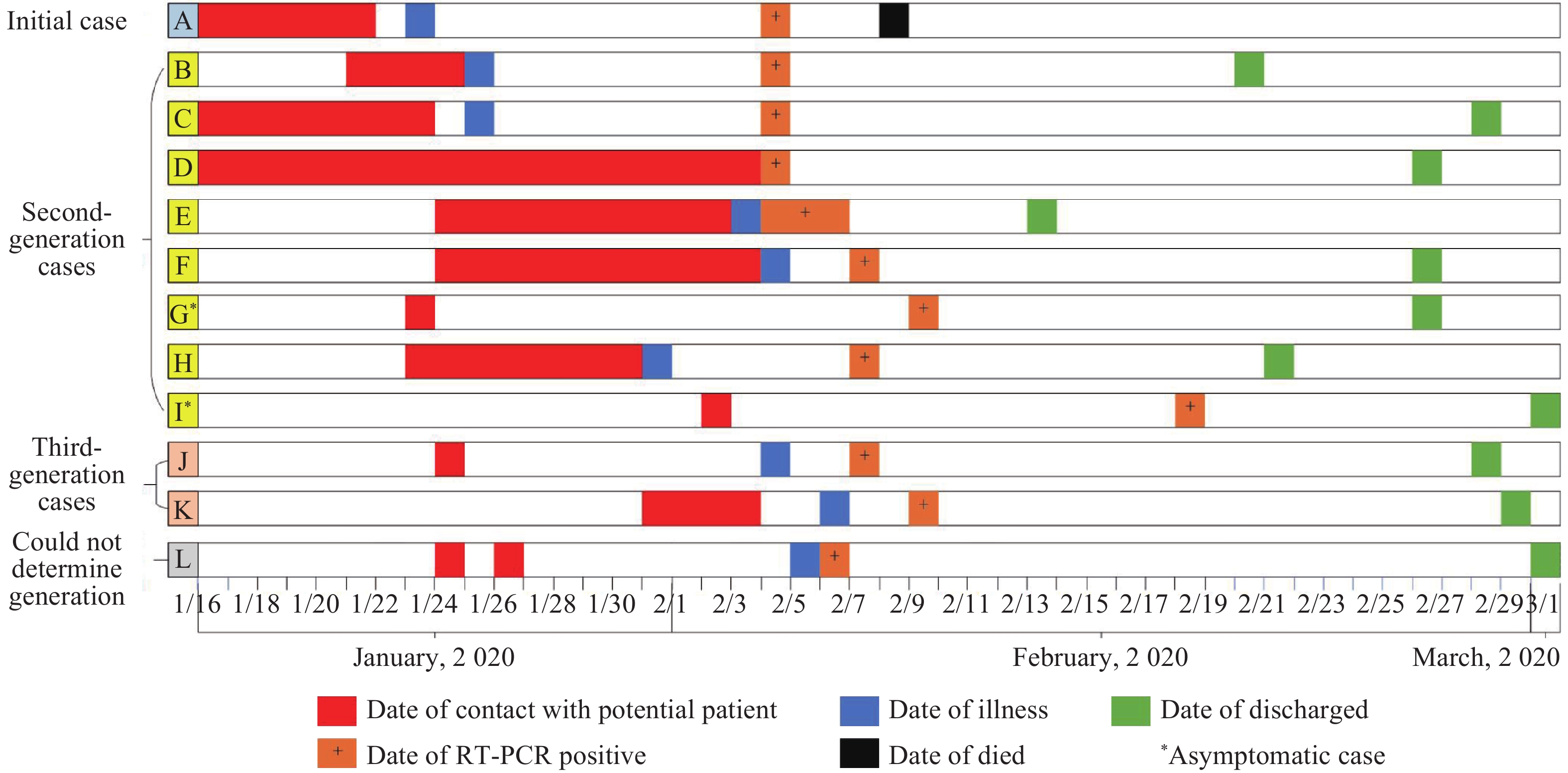
On February 2, 2020, a 28-year-old man (patient B) was isolated and treated in a designated hospital based on his high body temperature (39 ℃) during a visit to his grandmother (Patient A), who was an inpatient in this hospital. On February 4, COVID-19 was confirmed for both patient B and Patient A.
Patient A, an 85-year-old woman with multiple underlying diseases including coronary heart disease, hypertension, diabetes, and pulmonary interstitial fibrosis, was the only patient who indicated that she had been in contact with people from Hubei Province (the suspected origin of COVID-19 within China). She had been hospitalized due to repeated pulmonary interstitial fibrosis since January 2, 2020. On January 23, she presented symptoms of shortness of breath, of fever (37.8 ℃) on January 26, and of chest distress, dyspnea, and cough on January 30. She was classified as a critical case beginning on February 3 and died on February 8. Epidemiological investigations revealed that between January 16 and 21, 2 individuals from Hubei Province went to this hospital to visit their family member, a trauma patient in the same ward as Patient A, and all 3 individuals developed no symptoms. Asymptomatic carrier transmission had been reported for COVID-19 (4), and Patient A was possibly infected by asymptomatic carriers from Hubei Province. To exclude the possibility of Patient A becoming infected through close contact with unknown infected persons, such as the asymptomatic carriers in the hospital, we sampled all 674 persons (including 497 staff members, 177 inpatients, and their family members) who were not classified as close contacts of Patient A and who had been in the hospital from 14 days before the onset of Patient A’s symptoms. These persons were tested from February 21 and February 23 for PCR test and serological test, and all were negative for COVID-19 nucleic acids and antibodies. However, we could not determine whether these individuals from Hubei were asymptomatic carriers because they returned to Hubei Province on January 21, but our evidence suggests that these individuals are the most likely source of transmission.
A total of 44 close contacts had contact history with Patient A without appropriate personal protective measures and were traced and sampled every 2 days. These close contacts included 30 medical workers, 10 patients and visitors in the same ward, and 4 family members of Patient A. Overall, 8 confirmed and asymptomatic cases (as of the date of testing) were identified between February 4 and 18 among these close contacts (Patients B–I; overall attack rate: 18.2%) including 2 medical workers (attack rate: 6.7%), 4 patients and visitors in the same ward (attack rate: 40.0%), and 2 family members (attack rate: 50%) of Patient A. Local CDCs traced and tested close contacts of Patients B–I, and on February 7 and 9, 2 confirmed cases (Patients J and K) were detected.
Overall, 12 (Patients A–L) COVID-19 confirmed and asymptomatic patients were identified in this cluster (Figure 1 and Table 1) and none of them had a history of travel outside Qingdao 14 days before the onset of illness. The incubation period ranged from 1 to 11 days. Patient A was defined as having the initial case; among the 11 secondary cases (Patients B–L), 8 (Patients B–I) were second-generation cases and were likely infected from Patient A; 2 (Patients J and K) were third-generation cases and were infected from Patients B and C; and 1 (Patient L) case had an undetermined generation.
Patient A B C D E F G H I J K L Relationship Grandma of patient B Grandson of Patient A Daughter of Patient A Same ward as Patient A Daughter of Patient D Daughter of Patient D Nurse of Patient A Adjacent ward of Patient A Doctor of Patient A Brother of Patient C Same ward with Patient C Lab physician of Patient A Age (years) 85 28 60 70 45 43 22 50 38 59 46 48 Sex Female Male Female Female Female Female Female Female Male Male Female Male Occupation Retired Work at home Retired Retired Housework Pharmacist Medical staff Patient care Medical staff Housework Office worker Medical staff Underlying disease Coronary heart disease, hypertension, diabetes, pulmonary interstitial fibrosis − Myocarditis Diabetes − Hypertension − − − Diabetes − Hypertension Note: “-” represents without underlying disease. Table 1. Demographic features of the cluster infected with COVID-19 in a hospital of Qingdao, 2020.
Based on the dates of illness onset of 5 pairs of cases in this cluster, we fitted a gamma distribution by using data from field investigations to estimate the serial interval distribution and estimated that the serial interval distribution had a mean of 7.0 ± 4.4 days. A total of 11 cases occurred in the hospital and only 1 case (Patient J) occurred during a family dinner: Patient J was a 59-year-old man who joined a family dinner with Patient B and Patient C on January 24, and he experienced a runny nose on February 4 and tested positive for COVID-19 on February 7. Patient J had no contact history with other suspected cases 14 days before illness onset. Patients B and C did not have symptoms until January 25, so the source of infection for Patient J was most likely Patients B and C during their incubation period. A total of 5 persons participated in this family dinner, and only patient J developed disease so the attack rate was 20%.
Of the 3 medical workers (Patients G, I, and L) that were infected (Table 1, Figure 1), 2 (Patients G and I) had a history of direct contact with Patient A and were an attending doctor and nurse of Patient A. Patient G performed nursing services for Patient A on January 24 and was laboratory-confirmed for COVID-19 on February 8 (without symptoms as of the test date). Patient I carried out nasal catheterization for Patient A on February 2, and he developed suspected positive result for COVID-19 nucleic acid on February 13 and was laboratory-confirmed on February 18. Patients G and I wore disposable surgical masks, caps, and isolation gowns but did not wear medical gloves when in contact with Patient A. Patient L was a doctor in the hospital laboratory and had no contact history with Patient A and other patients in the hospital, and most likely became infected through performing two routine blood examinations for Patient A (without contact with samples of other confirmed cases) in the hospital laboratory with a primary level of personal protection, including disposable surgical mask, latex gloves, disposable medical cap, and gown. However, we cannot exclude the possibility that Patient L became infected through indirect contact with other COVID-19 cases in this cluster (5) based on the existence of confirmed and asymptomatic cases in this hospital. No COVID-19 cases had been reported before this cluster in this hospital, and all 4 family members and neighbors of Patient L tested negative for COVID-19 RNA.
-
Based on the findings from this study, a superspreading event of COVID-19 associated with a hospital occurred in Qingdao City with multiple populations experiencing the risk of infection. In addition, people who are pre-symptomatic can transmit COVID-19 virus and asymptomatic carriers may also transmit the disease. Targeted control measures include rapid identification, diagnosis, and isolation of all potentially infected patients, including a high index of suspicion for transmissible diseases and implementation of universal infection control procedures in all areas of the healthcare facilities.
Acknowledgment: We acknowledge staff members of the county-level CDCs of Qingdao for their assistance in the field investigation and data collection.
Conflict of interest: No conflicts of interest were reported.
| Citation: |




 Download:
Download:




