2022 Vol. 4, No. 2
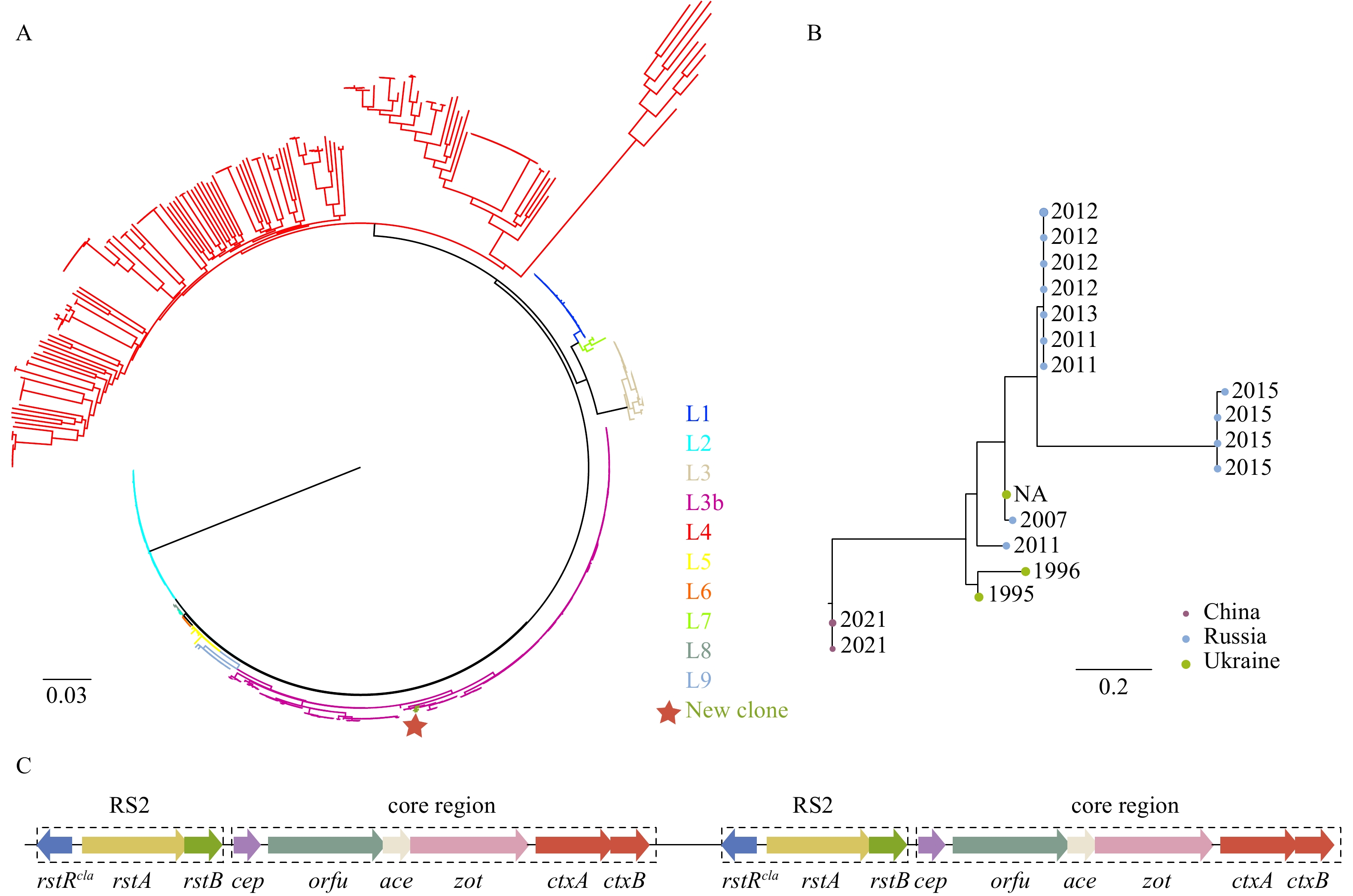
For a long time, Enterococcus faecium (E. faecium) was thought to be a commensal strain in human and animal digestive tracts. However, over the past three decades, some unique E. faecium clones rapidly acquired multiple antimicrobial resistance (AMR), which led these clones to survive hospital environments and become a hospital-adapted E. faecium clonal complex (CC) 17. Since the adaptation of these clones to changes in habitat, vancomycin-resistant E. faecium CC17 has emerged as the leading cause of hospital-acquired infections worldwide. This epidemic hospital-adapted lineage has diverged from other populations approximately 75 years ago. The CC17 lineage originated from animal strains, but not human commensal lines. We reviewed the evolutionary progress and the molecular mechanisms of E. faecium CC17 from a gut commensal to a multi-antimicrobial resistant nosocomial pathogen.
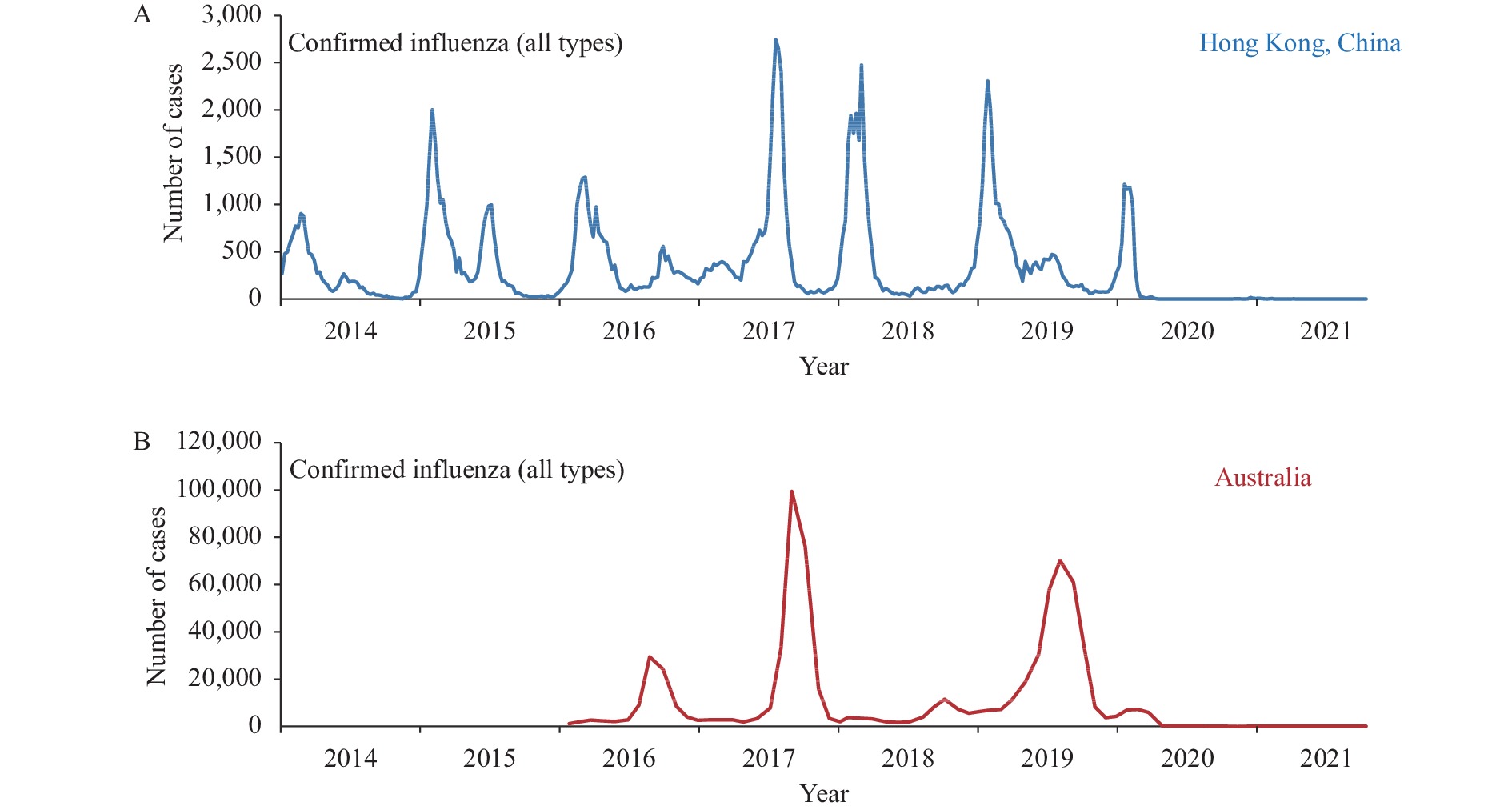
GII.2[P16] noroviruses (NoV) reemerged and rapidly became the main epidemic strain in acute gastroenteritis (AGE) outbreaks in Asian countries since 2016. The current GII.2 [P16] NoV showed the same antigenicity to the ones before 2016, but several unique amino acid substitutions existed in the RNA dependent RNA polymerase (RdRp) and other non-structural proteins, and the viral load of the current GII.2[P16] NoV was higher than those of other genotypes, it was estimated that the viral replication ability may have improved. However, other genotypes, such as GII.1 and GII.3, also had recombination with the novel RdRp, were not prevalent in AGE-outbreaks; thus, it was inferred that the capsid proteins also played an important role in the enhanced replication process. The viral infection could also be affected by other factors, such as the population genetic background, the climate and environment, and people’s lifestyles. Continued surveillance on genetic diversity and evolutionary pattern for the GII.2[P16] NoV is necessary.



 Subscribe for E-mail Alerts
Subscribe for E-mail Alerts CCDC Weekly RSS Feed
CCDC Weekly RSS Feed