2021 Vol. 3, No. 46
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a recently emergent coronavirus of natural origin and caused the coronavirus disease (COVID-19) pandemic. The study of its natural origin and host range is of particular importance for source tracing, monitoring of this virus, and prevention of recurrent infections. One major approach is to test the binding ability of the viral receptor gene ACE2 from various hosts to SARS-CoV-2 spike protein, but it is time-consuming and labor-intensive to cover a large collection of species.
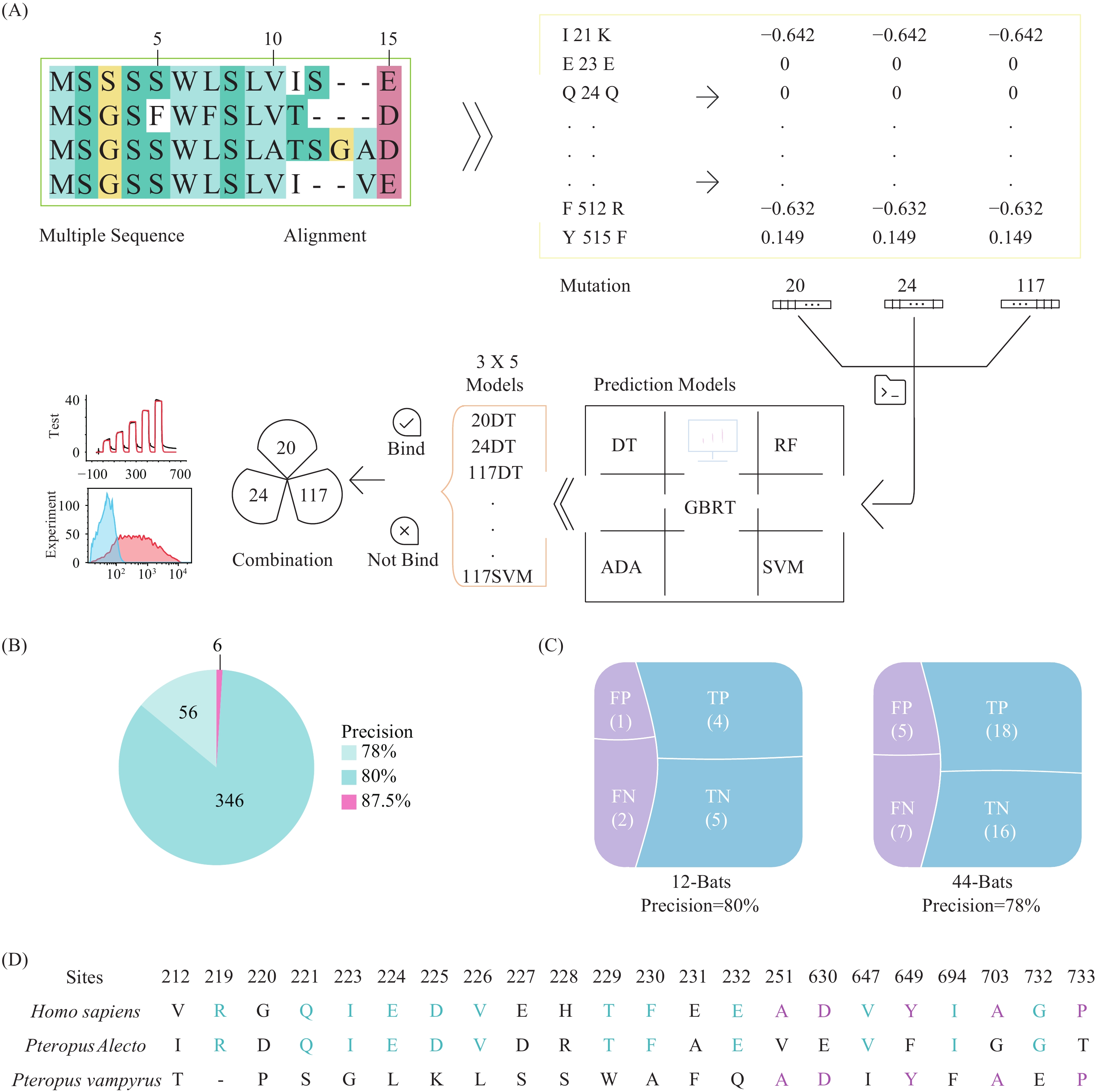
In this paper, we applied state-of-the-art machine learning approaches and created a pipeline reaching >87% accuracy in predicting binding between different ACE2 and SARS-CoV-2 spike.
We further validated our prediction pipeline using 2 independent test sets involving >50 bat species and achieved >78% accuracy. A large-scale screening of 204 mammal species revealed 144 species (or 61%) were susceptible to SARS-CoV-2 infections, highlighting the importance of intensive monitoring and studies in mammalian species.
In short, our study employed machine learning models to create an important tool for predicting potential hosts of SARS-CoV-2 and achieved the highest precision to our knowledge in experimental validation. This study also predicted that a wide range of mammals were capable of being infected by SARS-CoV-2.
The best approach to preventing the importation of coronavirus disease 2019 (COVID-19) is enhancing the detection capacity at customs. The rapid detection is of utmost importance and therefore highly demanded.
We conducted a field validation study of a duplex real-time reverse transcription recombinase-aided amplification (RT-RAA) assay in Zhoushan and Hangzhou customs, in Zhejiang Province, China. The reverse transcriptase polymerase chain reaction (RT-PCR) assay kit routinely used at customs was used in parallel, and the duration the two methods took to complete a specific number of samples was compared.
Among 506 samples collected, RT-RAA results were consistent with the RT-PCR results. The sensitivity and specificity were 100%, the total coincidence rate was 100%, and the Kappa value was 1 (P<0.05) for both methods. The RT-RAA kit took a significantly shorter time in testing the 20–200 samples than the RT-PCR kit.
The RT-RAA detection method is more efficient and suitable for use at customs than RT-PCR assay to realize rapid customs clearance of 200 or fewer samples.



 Subscribe for E-mail Alerts
Subscribe for E-mail Alerts CCDC Weekly RSS Feed
CCDC Weekly RSS Feed