2020 Vol. 2, No. 25
What is already known on this topic?
Coronavirus disease 2019 (COVID-19), a disease caused by a novel human coronavirus named the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) or COVID-19 virus, was reported in December 2019. Complete genomes of the COVID-19 virus from clinical samples using next generation sequencing (NGS) have been reported.
What is added by this report?
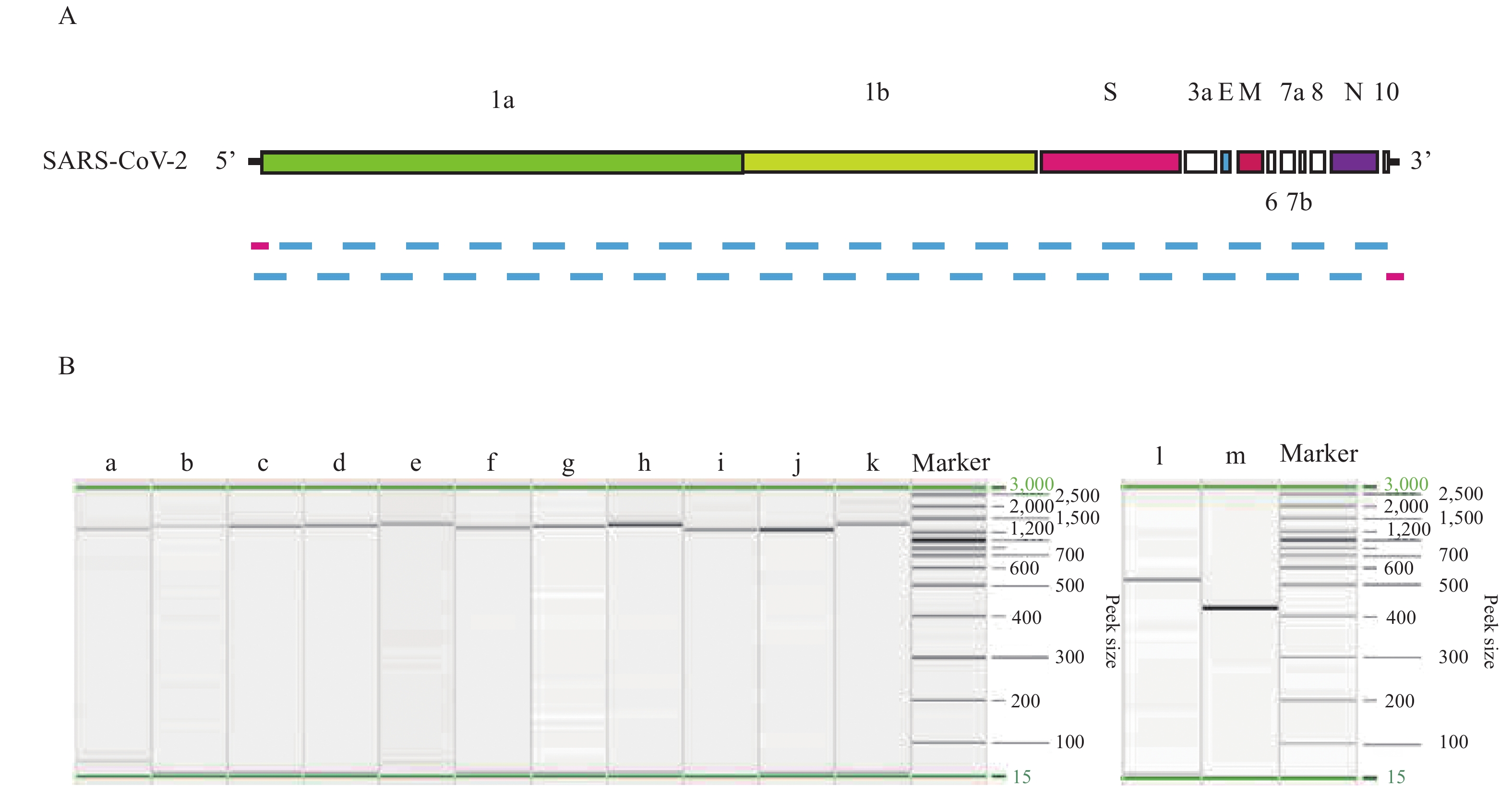
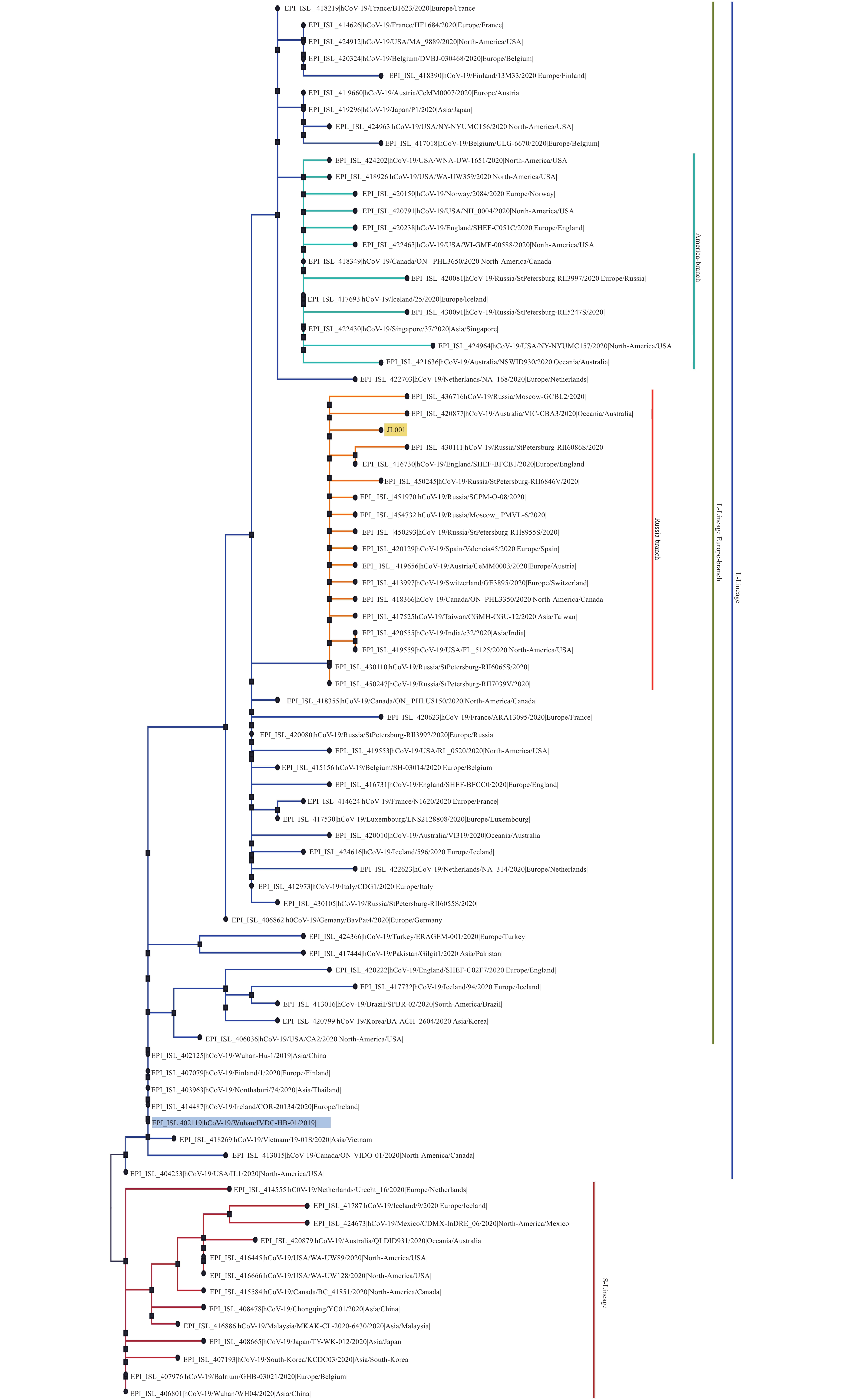
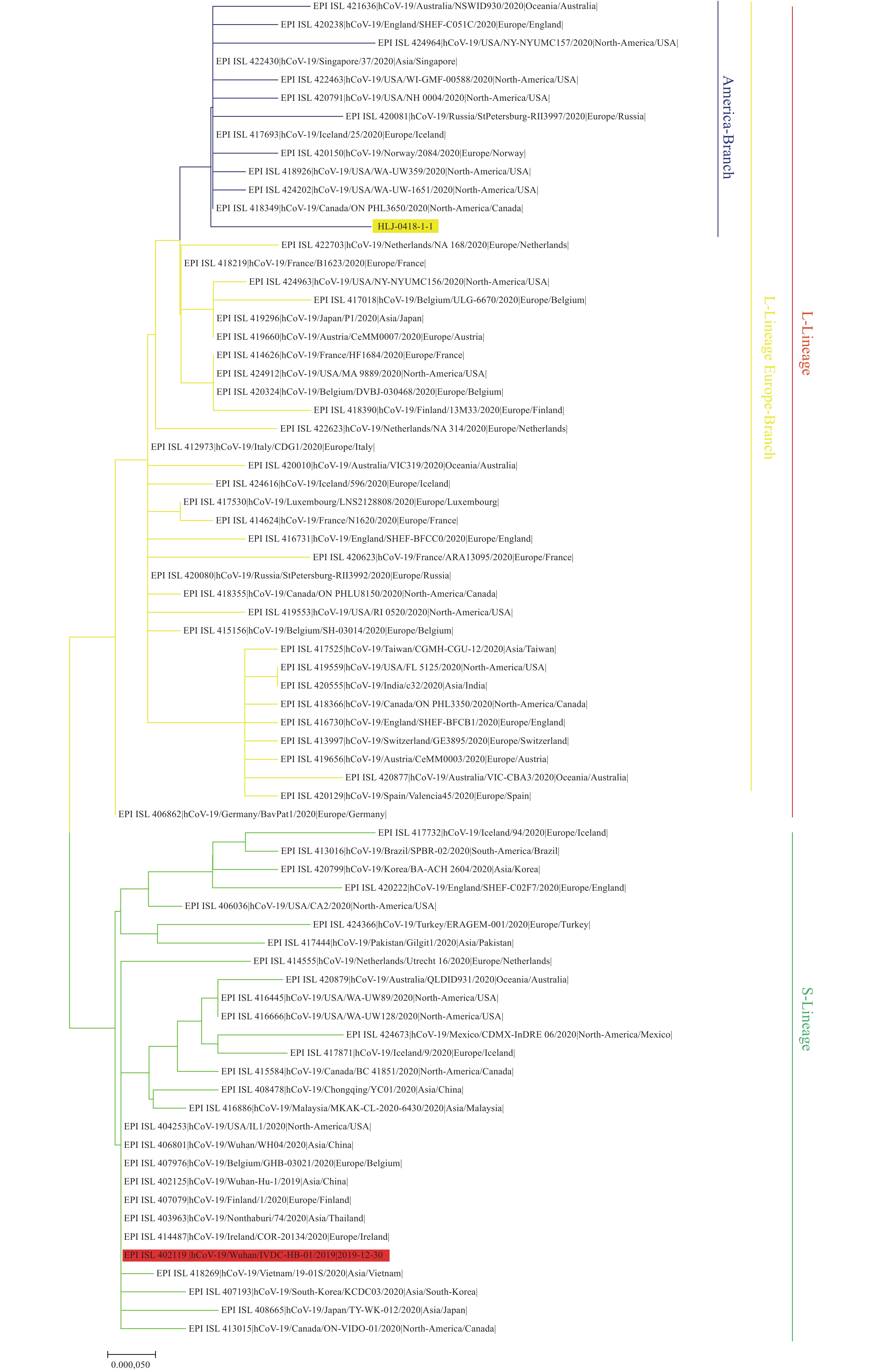
Here we provide the technical data for sequencing complete genome of COVID-19 virus from clinical samples using the Sanger method. Two complete COVID-19 virus genome sequences (named WH19004-S and GX0002) were obtained from clinical samples of COVID-19 patients, and two single nucleotide polymorphisms (SNPs) in ORF7a (T/C, nt 27,493) and ORF8 (T/C, nt 28,253) of WH19004-S were identified by Sanger sequencing.
What are the implications for public health practice?
The COVID-19 virus genome sequencing by Sanger method reported here could be used to generate data of high enough quality without requirement for expensive NGS equipment, which support sequencing complete genomes from clinical samples and monitoring of viral genetic variations of COVID-19 infections.
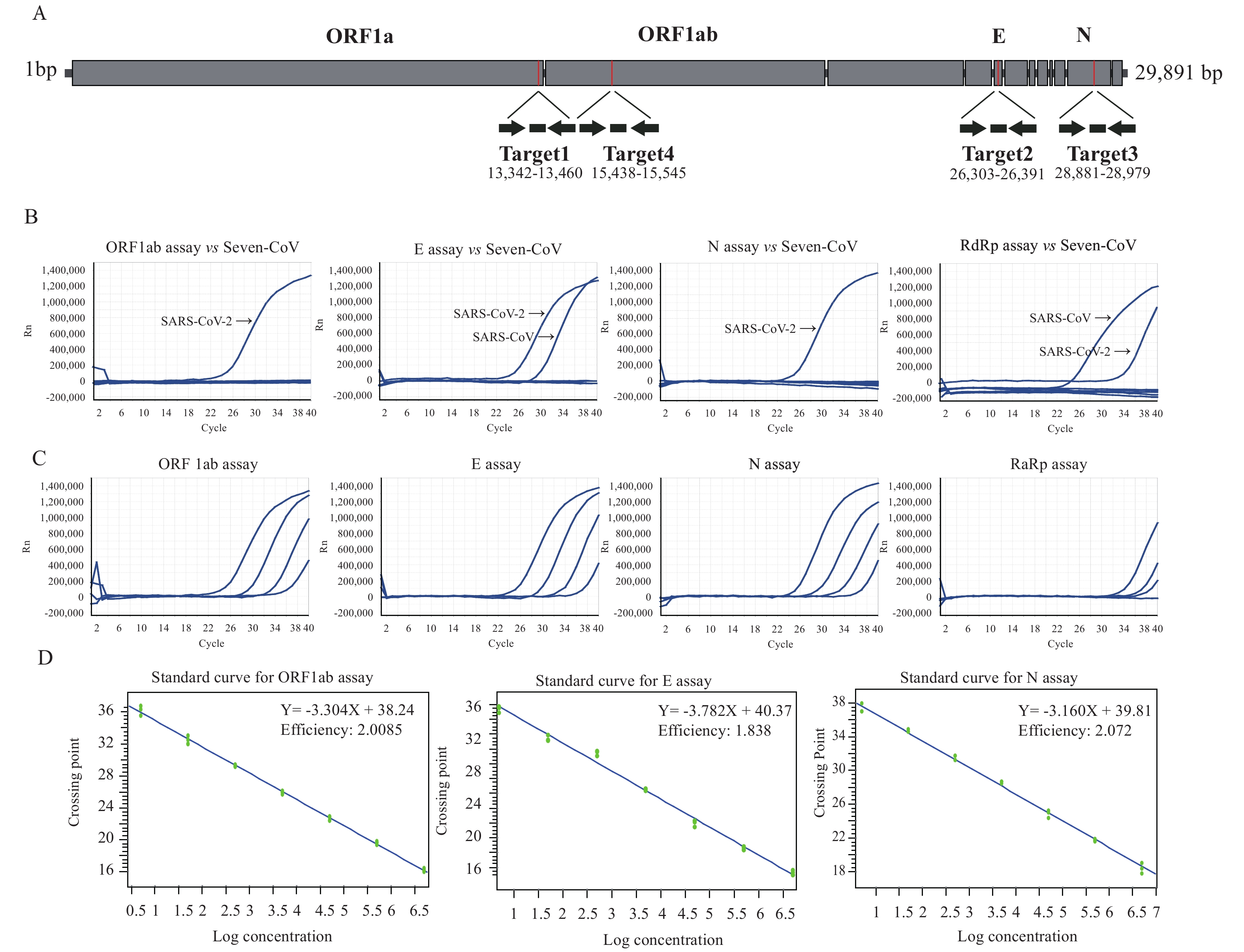
A novel human coronavirus, known as SARS-CoV-2 or 2019-nCoV, is the causative agent of the coronavirus disease 2019 (COVID-19). We have released the primers and probes of real-time reverse transcription polymerase chain reaction (rRT-PCR) assays for the laboratory detection of COVID-19 infection.
Here we provide detailed technical data and evaluate the performance of three novel rRT-PCR assays targeting the ORF1ab, N, and E genes for detection of COVID-19 infection. The application of rRT-PCR assays among four types of specimens (alveolar lavage, sputum, throat swabs, and stool) from patients with COVID-19 indicated that the mean viral loads detected in sputum were higher than other specimens.
These rRT-PCR assays reported here could be used for laboratory diagnosis of COVID-19 infection with high sensitivity, specificity, and applicability. Sputum rather than throat swabs and stool should be a priority for specimen collection for laboratory detection of COVID-19.



 Subscribe for E-mail Alerts
Subscribe for E-mail Alerts CCDC Weekly RSS Feed
CCDC Weekly RSS Feed