-
Coronaviruses are single-stranded positive-stranded RNA that have the largest virus genome among RNA viruses (1-2). Coronaviruses are widespread in bats around the world but can be found in many other species as well that are phenotypically and genotypically diverse (1,3). The coronavirus disease 2019 (COVID-19) raised considerable concerns as it was associated with severe acute pneumonia and fatal outcomes(4-5) and thus resembled the clinical presentation of severe acute respiratory syndrome (SARS) observed in 2002 and 2003 as well as Middle East respiratory syndrome (MERS) since 2012 (6). The causative agent of COVID-19 was a novel coronavirus known as SARS-CoV-2 and was previously named 2019-nCoV in China (2,5).
Novel quantitative real-time reverse-transcription polymerase chain reaction (rRT-PCR) assays were developed in rapid response to the emergence of COVID-19 originating in Wuhan, and these assays have been widely used in laboratory detection and written into the national technical guidelines used in China (7). We report here in detail on the technical data and comparative analysis of performance of three rRT-PCR assays targeting three distinct regions of the SARS-CoV-2 genome for detection of COVID-19 infection. Three rRT-PCR assays were further evaluation with several species of clinical specimens from patients with COVID-19.
HTML
-
A total of 135 clinical specimens were collected from a cluster of patients with pneumonia in Wuhan and Beijing suspected of being infected with SARS-CoV-2. Specimens included alveolar lavage, sputum, throat swabs, and stool. Inactivation of specimen processing was performed in a biosafety level 3 (BSL3) laboratory.
-
Nucleic acids were extracted from a 140 μL processed specimen using a QIAamp Viral RNA Mini Kit according to the manufacturer’s instructions. Approximately 60 μL of total nucleic acid eluate was recovered into nuclease-free tubes and either tested immediately or stored at -70 °C.
-
Multiple primer and probe sets were designed based on bioinformatics analysis of three complete genomes of SARS-CoV-2 (BetaCoV/Wuhan/IVDC-HB-01/2019, Accession ID: EPI_ISL_402119, BetaCoV/Wuhan/IVDC-HB-04/2020, Accession ID: EPI_ISL_402120) obtained in our lab (5,8). ORF1ab, E gene and N gene sequences were selected as targets using Primer Premier software version 5 (Applied Biosystems) with the following default settings: primer melting temperature (TM) set at 60 °C; probe TM set at 10 °C greater than the primers at approximately 70 °C; and no guanidine residues permitted at the 5’ probe termini (Figure 1A, Table 1). Previous reported prime and probe sets for the RNA-dependent RNA polymerase (RdRp) region of pan beta-CoV was designed as a reference experiment (3,9).
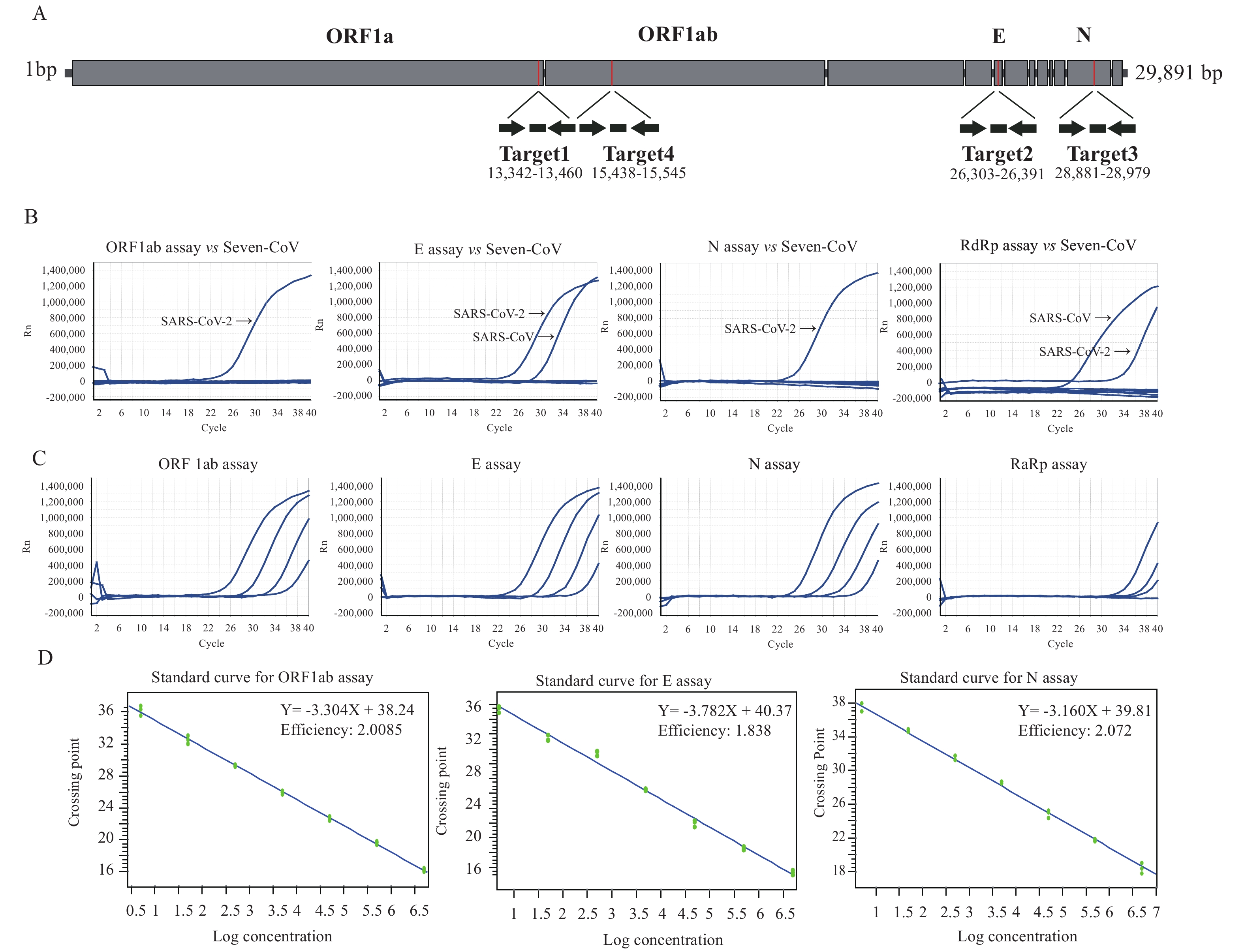 Figure 1.
Figure 1.Development of three rRT-PCR assays for detection of SARS-CoV-2. (A) Description of specific primers and probes for detection of SARS-CoV-2. Relative positions of amplicon targets on the SARS-CoV-2 genome schematic. ORF: open reading frame; RdRp: RNA-dependent RNA polymerase gene; E: envelope protein gene; N: nucleocapsid protein gene. Numbers below amplicons are genome positions according to SARS-CoV-2, accession ID: EPI_ISL_402119, EPI_ISL_402120. (B) Confirmation of rRT-PCR on seven human coronavirus samples using specific probes and primers. (C) Representative amplification plot of developed rRT-PCR assay showing serial dilutions of SARS-CoV-2 RNAs from clinical samples for evaluation of sensitivity of four assays. (D) Determination of detection efficiency and limit of various rRT-PCR assays. Standard calibration curves of Ct and genomic copy number was generated based on rRT-PCR results targeting primer set and SARS-CoV-2 stock with pre-determined genomic copies.
Target Gene Primer Sequence (5’→3’) Genome location* Limit of detection (copies/mL) Forward CCCTGTGGGTTTTACACTTAA Target 1 ORF1ab Reverse ACGATTGTGCATCAGCTGA 13342-13460 203 Probe FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1 Forward TTCTTGCTTTCGTGGTATTC Target 2 E Reverse CACGTTAACAATATTGCAGC 26303-26391 664 Probe FAM-GTTACACTAGCCATCCTTACTGCGCTTCGA-BHQ1 Forward GGGGAACTTCTCCTGCTAGAAT Target 3 N Reverse CAGACATTTTGCTCTCAAGCTG 28881-28979 667 Probe FAM-TTGCTGCTGCTTGACAGATT-BHQ1 Forward GGTCATGTGTGGCGGCTC Target 4 RdRp Reverse GCTGTAACAGCTTGACAAATGAAAG 15438-15545 Probe FAM-CTATATGTTAAACCAGGTGGAAC-BHQ1 *The location on the reference genome, accession ID: EPI_ISL_402119, EPI_ISL_402120. Table 1. Primer and probe sets of rRT-PCR assays for the detection of the 2019 novel coronavirus diseases (COVID-19).
-
Several one step rRT-PCR assays were developed using the OneStep PrimeScript™ RT-PCR kit (TaKaRa, Japan). Each 25 µL reaction mixture contained 12.5 µL of 2× Master Mix, 0.5 µL of reverse transcriptase/Taq DNA polymerase mixture, 5 μL of RNA extract, 400 nmol/L concentrations of forward primer and reverse primer, and 200 nmol/L of probe. Thermal cycling included 42 °C for 5 minutes, followed by 95 °C for 10 seconds and then 40 cycles of 95 °C for 10 s and 60 °C for 45 s. Each run included one SARS-CoV-2 genomic template control and at least two negative or mock controls (for the extraction and the PCR amplification step) (Table 1).
Assay specificity was determined using high-titer virus stock as well as clinical samples containing known respiratory viruses from collection of our laboratory. Identities and virus RNA concentrations were reconfirmed by specific rRT-PCRs for each virus before the experiment (9-10).
The limit of detection was independently assessed using a SARS-CoV-2 stock with pre-determined genomic copies. The calibration curve for the genomic copy number versus Ct value was obtained from the rRT-PCR machine, which was an ABI 7500 (Applied Biosystems, USA) or a Roche LightCycler 480 (Roche, Switzerland) Real-Time PCR system. Series of four parallel reactions per concentration step were prepared and tested by the respective rRT-PCR.
Clinical samples were considered positive if two or more of the gene targets showed positive results (Ct≤37 cycles; If the Ct value is between 37–40, the experiment needs to be repeated. The judgment of the final result is based on two consistent experimental results) (7).
Clinical Specimens
Nucleic Acid Extraction
Design of Primers and Probes
Real-Time RT-PCR Assay for Screening of SARS-CoV-2 Infection
-
In the detection of clinical samples using the same gradient dilution, ten-fold serial dilutions of the SARS-CoV-2 RNA were tested to assess the detection limits and dynamic range of our optimized rRT-PCR assays. The detection results of the four rRT-PCR assays were different for clinical samples with gradient dilution. The lower potential limit of detection was approximately 10-4 dilution per reaction for ORF1ab assay, E assay, and N assay, and 10-3 dilution per reaction for RdRp assay (Figure 1C). The genomic copy numbers-based Ct values were calibrated on the standard curve for individual rRT-PCR (Figure 1D). The lowest detection limit of ORF1ab assay was 203 copies/mL. The minimum detection limits of E and N assay were 664 and 667 copies/mL, respectively (Table 1).
The rRT-PCR assays were tested with nucleic acid extracts of 6 human coronaviruses, human NL63 coronaviruses (NL63-CoV), human 229E coronaviruses (229E-CoV), human OC43 coronaviruses (OC43-CoV), human HKU1 coronaviruses (HKU1-CoV), SARS-CoV, and MERS-CoV. In addition, nucleic acid extracts of influenza A were also tested. No positive reactions were observed with any of the primer and probe sets of ORF1ab and N genes, while cross-reactions with SARS-CoV were shown using the primer and probe sets of the E gene as well as RdRp (Figure 1B).
Assay reproducibility was tested by using replicate ten-fold serial dilutions of the SARS-CoV-2 RNA and intra- and inter-assay variability evaluated for each dilution point in quadruplicate three different times. The reproducibility for 3 assays (targeting ORF1ab, E, and N) exceeded 90% at the lower copy detection limit (data not shown).
-
The rRT-PCR assay was evaluated using different species of clinical specimens collected from a cluster of patients in Wuhan and Beijing, whom were suspected of being infected with SARS-CoV-2. Three rRT-PCR targeting ORF1ab, N and E showed consistent detection rates among various species of specimens (Table 2). The mean Ct value detected in sputum was lower than in other species of clinical specimens (alveolar lavage, throat swab, and tool), which indicated the mean viral loads detected in sputum were higher than other specimens. Similar results were also reported recently using a large amount of clinical specimens.
Target gene Alveolar lavage n(%) Sputum n(%) Throat swab n(%) Stool n(%) Total n=15 n=28 n=53 n=39 ORF1ab Positive rate (%) 14 (93.33%) 12 (42.86%) 25 (47.17%) 16 (41.03%) Mean Ct value 31.11 28.44 32.53 29.93 Range 26.41−36.25 18.44−36.38 24.22−38.58 22.27−37.51 N Positive rate (%) 14 (93.33%) 12 (42.86%) 25 (47.17%) 16 (41.03%) Mean Ct value 31.45 28.91 33.61 31.14 Range 26.58−36.98 18.04−37.76 25.73−38.31 23.46−38.11 E Positive rate (%) 14 (93.33%) 12 (42.86%) 25 (47.17%) 16 (41.03%) Mean Ct value 31.33 28.64 33.27 30.46 Range 26.45−36.55 18.34−36.42 25.03−38.54 22.46−38.30 Table 2. Detection results of clinical specimen by the rRT-PCR based on molecular targets of ORF1ab, N and E gene.
Performance of Different rRT-PCR Methods
Evaluation with Different Species of Clinical Specimens
-
To date, several coronaviruses (including SARS-CoV, MERS-CoV, and SARS-CoV-2) that could infect humans have all been beta-coronaviruses (1,8). The risk associated with false-positive PCR results posed a challenge in clinical application. First, the primer dimers and non-specific amplification, in which probe sequences participate, might interfere with experiments. Second, SARS-CoV-2 assays might cross-react with other viruses. In this study, we used 6 human coronaviruses (NL63-CoV, 229E-CoV, OC43-CoV, HKU1-CoV, SARS-CoV and MERS-CoV) and influenza A to test the cross-reactivity. Consequentially, the ORF1ab and N gene-based assay were the most specific, exactly matching target genes of SARS-CoV-2. The E gene was cross-reacted with B lineages of the beta-coronavirus (such as SARS-CoV). We propose that the E gene could be used as a broad-spectrum screening gene for B lineages of the beta-coronavirus, such as SARS-CoV, SARS-CoV-2, bat SARS-like coronavirus, et al. Therefore, we currently prefer to recommend the ORF1ab and N gene-based rRT-PCR assays for detection of COVID-19 infection.
Here, we also verified the sensitivities and detection limits of various rRT-PCR assays. The data showed that similar detection rates and sensitivity were observed among three rRT-PCR assays for ORF1ab, E gene, and N gene targets, which were higher than the RdRp target. We were also noticed that the ORF1ab-based rRT-PCR assay showed the best sensitivity (lowest detection limit).
To determine which type of specimen is suitable for rRT-PCR in clinic, four species of clinical specimens (alveolar lavage, sputum, throat swab, and stool) were collected for evaluation. Our data indicated that the mean viral loads detected in sputum were higher than other specimens, which suggests that sputum should be a priority for collection for laboratory detection of COVID-19 (11-12).
Although an rRT-PCR offers clear advantages over more conventional RT-PCR formats, assay results must still be interpreted with caution. For example, the effectiveness of rRT-PCR for detection of SARS-CoV-2 in clinical specimens had been shown to be greatly influenced by the quantity, type, and timing of specimen collection (11-13). In addition, false-negative results due to poor quality nucleic acids or presence of rRT-PCR inhibitors could also be a concern. False-negative results could also potentially arise from mutations occurring in the primer and probe target regions of SARS-CoV-2 genome. We included multiple genetic targets in our assay based on analysis of SARS-CoV-2 genomes available. Finally, to avoid false-positive results, steps should be taken meticulously to prevent introduction of contaminating viral RNA or previously amplified DNA during preparation of nucleic acid extracts and amplification reactions.
Conflicts of interest: The authors declare that they have no conflict of interest.
Fundings: This work was supported by the National Key Research and Development Program of China (2016YFD0500301, 2020YFC0840900).
| Citation: |



 Download:
Download:




