-
At the end of 2019, coronavirus disease 2019 (COVID-19) caused by COVID-19 virus, also known as SARS-CoV-2, was first discovered and quickly began to spread around the world (1). Some researchers evaluated and compared the whole genome sequences of circulating COVID-19 and found mutations associated with the infectivity of the virus. High-frequency mutations in the COVID-19 genome were found in nsp6, RNA polymerase, helicase, membrane glycoprotein, RNA primer, nucleocapsid phosphoprotein, and spike protein genes (2). One of the most critical mutations was the D614G of the spike protein gene (S), which is a replacement of aspartic acid (D) with glycine (G). The transmission of S-G614 mutants was stronger than that of S-D614 mutants because a newly-formed serine protease called elastase-2 in the S-G614 mutant could lead to an increase in enzyme cleavage efficiency and infectivity (3–4).
The D614G mutation brings new challenges to the prevention and control of the epidemic, and there are few reported molecular detection methods for D614G mutation (5). Therefore, developing a detection method to distinguish S-G614 mutants from S-D614 mutants is highly important. Many conventional methods are available for detecting single nucleotide polymorphism (SNP), such as real-time PCR (6), DNA sequencing (7), restriction fragment length polymorphism (RFLP) (8). However, these methods are either time-consuming and laborious or require more sophisticated instruments and skillful personnel.
Isothermal DNA amplification technology offers a good alternative to mutation detection due to its simplicity and rapidity. Among them, loop-mediated isothermal amplification (LAMP) is the most common method for SNP detection, but the design of LAMP primers is complex, and its application is limited by the requirement of a high temperature (65 °C) (9). There are also some mutation detection studies on recombinase polymerase amplification (RPA) (10), its sensitivity and specificity are similar with PCR, but expensive (7.7 USD/test). Recombinase-aided amplification (RAA) is a new isothermal amplification assay for various pathogens (11) and uses specific enzymes and proteins to quickly detect nucleic acid in less than 30 minutes at 39 °C, and it is cheaper (4.6 USD/test). Recently, a modified RAA assay, probe-directed recombinase amplification (PDRA), was developed in our laboratory for detection of SNPs (12-13). In this study, a PDRA method for simple and quick differentiation of D614G mutation in COVID-19 was reported.
-
A total of 53 previously confirmed COVID-19 virus positive RNAs and 10 negative RNAs (from other respiratory viral pathogen-positive swab samples) were all stored in the National Institute for Viral Disease Control and Prevention of China CDC in Beijing, China.
-
The QIAGEN OneStep RT-PCR Kit for PCR and sequencing were used for comparison. The PCR forward primer sequence was 5-AATCTATCAGGCCGGTAGCAC-3. The reverse sequence was 5-CACCAATGGGTATGTCACACT-3. (14) The PCR assay was performed in a 25 μL reaction system containing 5 μL of 5×PCR buffer, 2 μL of One Step RT-PCR Enzyme Mix (QIAGEN), 1 μL dNTP mix, 0.1 μL of RNase inhibitor (RRI), 2 μL of primer mix (10 μmol/L), and 2 μL of template nucleic acid using a CFX96 Real-Time PCR System (Bio-Rad, USA) under the following conditions: a 30 min reverse transcription step at 50 °C, a 15 min denaturation step at 95 °C, and followed by 35 cycles at 94 °C for 30 s, 55 °C for 1 min, and 72 °C for 1 min, a 10 min final extension step at 72 °C, and finally kept at 4 °C. The PCR products were then sent to Sangon Biotech (Shanghai, China) for sequencing.
-
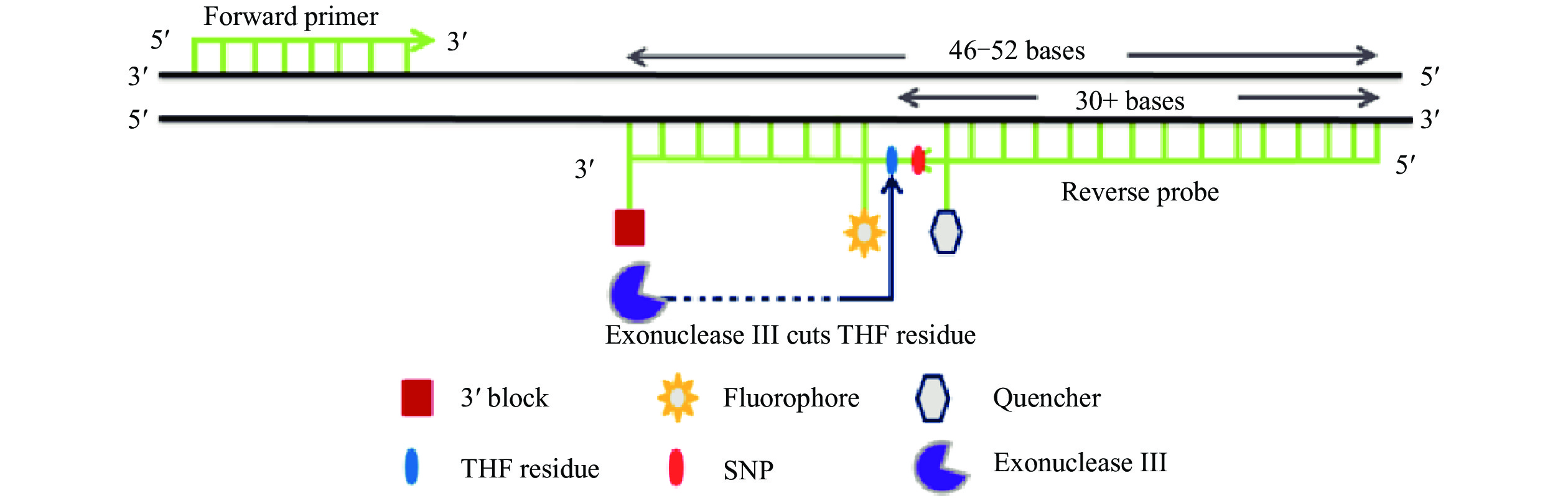
The PDRA assay included two real-time RAA reactions (A and G) to detect the A and G nucleotides of the D614G mutation (A23403G polymorphism), respectively. Each individual PDRA reaction contained a primer and a probe. The forward primers of the two reactions were the same, and there was only one nucleotide difference in the polymorphic site between the two probes. the primers and probe design were based on the S protein sequence of SARS-CoV-2 with accession number MT252819.1. A forward primer and two specific reverse probes for A and G nucleotides of A23403G polymorphism were designed using Oligo7 software. The primer is usually 30–35 bp in length and the probe is usually 46–52 nucleotides in length, of which at least 30 nt is located at the 5’ end, adjacent to the internal base-free site mimic (tetrahydrofuran [THF]) site, flanked by dT-fluorophore and the corresponding dT-quencher group, and at least 3 nucleotides at the 3’ end. At the same time, the mutation site was designed to be just before the THF site of each probe (Figure 1). The forward primer sequence was 5-TTGAGATTCTTGACATTACACCATGTTCTTT-3. The reverse probe D sequence was 5-TAGCAACAGGGACTTCTGTGCAGTTAACATCCTGATAAAGAACAGC-3. The reverse probe G sequence was 5-TAGCAACAGGGACTTCTGTGCAGTTAACACCCTGATAAAGAACAGC-3. All the primers and probes were synthesized and purified by Sangon Biotech (Shanghai, China) using high-performance liquid chromatography (HPLC).
-
PDRA assay was performed using RAA kits (Jiangsu Province Qitian Biology Co., Ltd., China), and the reaction system was partially modified. The A and G reactions were divided into two tubes (A and G). A reaction was carried out in a 50 μL lyophilized reaction tube containing a dried enzyme precipitation 0.2 mL, 25 μL rehydration buffer, 18.2 μL ddH2O, 1.3 μL forward primers (10 μmol/L), 1 μL A nucleotide specific PDRA probe (10 μmol/L), 2 μL DNA template, and 2.5 μL magnesium acetate (280 mmol/L). G reaction was carried out in a 50 μL lyophilized reaction tube containing a dried enzyme precipitation 0.2 mL, 25 μL rehydration buffer, 17.9 μL ddH2O, 1.6 μL forward primers (10 μmol/L), 1 μL G nucleotide specific PDRA probe (10 μmol/L), 2 μL DNA template, and 2.5 μL magnesium acetate (280 mmol/L). The test tube was then transferred to the test tube rack of RAA fluorescence detection equipment QT-RAA-F7200 (Jiangsu Province Qitian Biology Co., Ltd., China) and incubated at 39 °C for 30 min. Negative controls (water without nuclease) were included in each run.
-
When the slope of the fluorescence curve is >20, it is judged to be a positive result by the detector, and when the positive result of probe D appears earlier than that of probe G and the time difference ≥3 min, it is interpreted as S-D614 strain. When the positive result of probe G appeared earlier than that of probe D and the time difference ≥3 min, it was interpreted as S-G614 mutant.
-
The recombinant plasmids of S-G614 mutant and S-D614 strain with a concentration in the 10-fold range of 102 copies/μL to 107 copies/μL were used to validate the sensitivity and specificity of PDRA test. In different 5 days, 20 repeated experiments were carried out to verify its repeatability.
-
PDRA was used to detect 53 COVID-19 positive nucleic acid samples and 10 negative samples the results were compared with those of the sequencing results.
-
The sensitivity of PDRA for the detection of two recombinant plasmids was 103 copies (Figure 2) and the reproducibility was shown in Table 1. No cross reaction between two recombinant plasmids (G and D) was observed within at least 3-minute interval by corresponding probes in the detection range (Figure 3). A total of 63 clinical samples were typed by PDRA and direct sequencing. The sequencing results of 63 clinical samples were compared with the results of PDRA (Figure 4), and the coincidence rate was 100% (Table 2). Among the 63 clinical samples, there were 22 S-D614 mutants, 31 S-G614 mutants, and 10 negative samples.
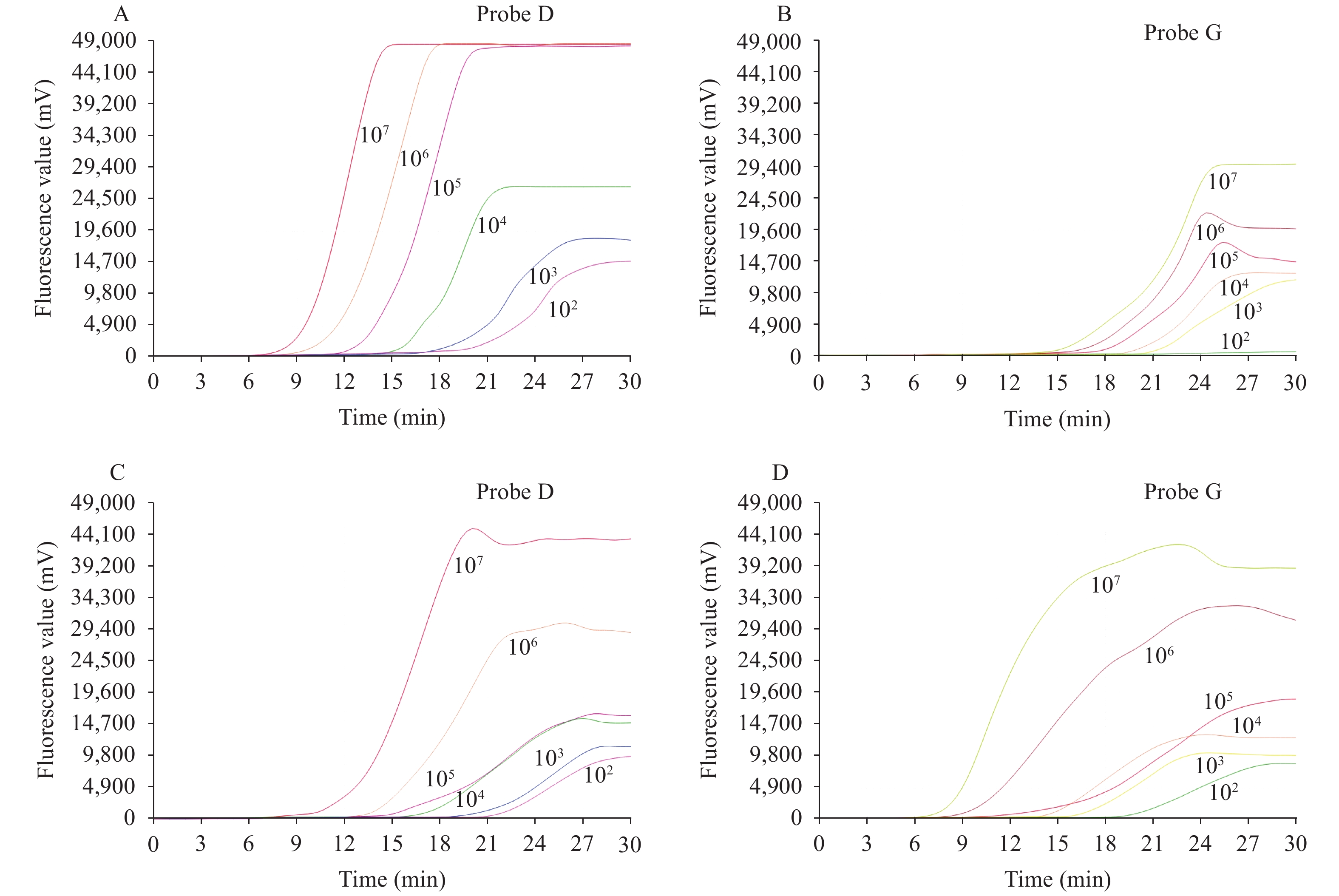 Figure 2.
Figure 2.Sensitivity result of PDRA with recombinant plasmids. (A) Probe D tested S-D614 mutant recombinant plasmids; (B) Probe G tested S-D614 mutant recombinant plasmid; (C) Probe D tested S-G614 mutant recombinant plasmids; (D) Probe G tested S-G614 mutant recombinant plasmids.
Note: When the S-D614 strain recombinant plasmid was detected, the sensitivity of Probe D and Probe G was 102 copies/reaction and 103 copies/reaction, respectively, and the time to positivity by probe D at each concentration was less (at least 3 min interval) than that by probe G. When the S-G614 mutant recombinant plasmid was detected, the sensitivity of both Probe D and Probe G was 102 copies/reaction. The time to positivity by probe G at each concentration was less (at least 3 min interval) than that by probe D.Serial diluted DNA No. of replicates tested No. detected Detection rate (%) 107 20 20 100 106 20 20 100 105 20 20 100 104 20 20 100 103 20 20 100 102 20 2 10 Table 1. The reproducibility of probe-directed recombinase amplification (PDRA) assay.
 Figure 3.
Figure 3.Specificity result of PDRA with recombinant plasmids. (A) S-D614 strain recombinant plasmid; (B) S-G614 mutant recombinant plasmid.
Note: When the 107 copies of S-D614 strain recombinant plasmid were detected, the time to positivity by Probe D was less than that of Probe G by 6 min. When the 103 copies of recombinant plasmid were detected, the time to positivity by Probe D was less than that of Probe G by 4 min. When the 107 copies and 103 copies of S-G614 mutant recombinant plasmids were detected, the time to positivity by Probe G was less than that of Probe D by 3 min.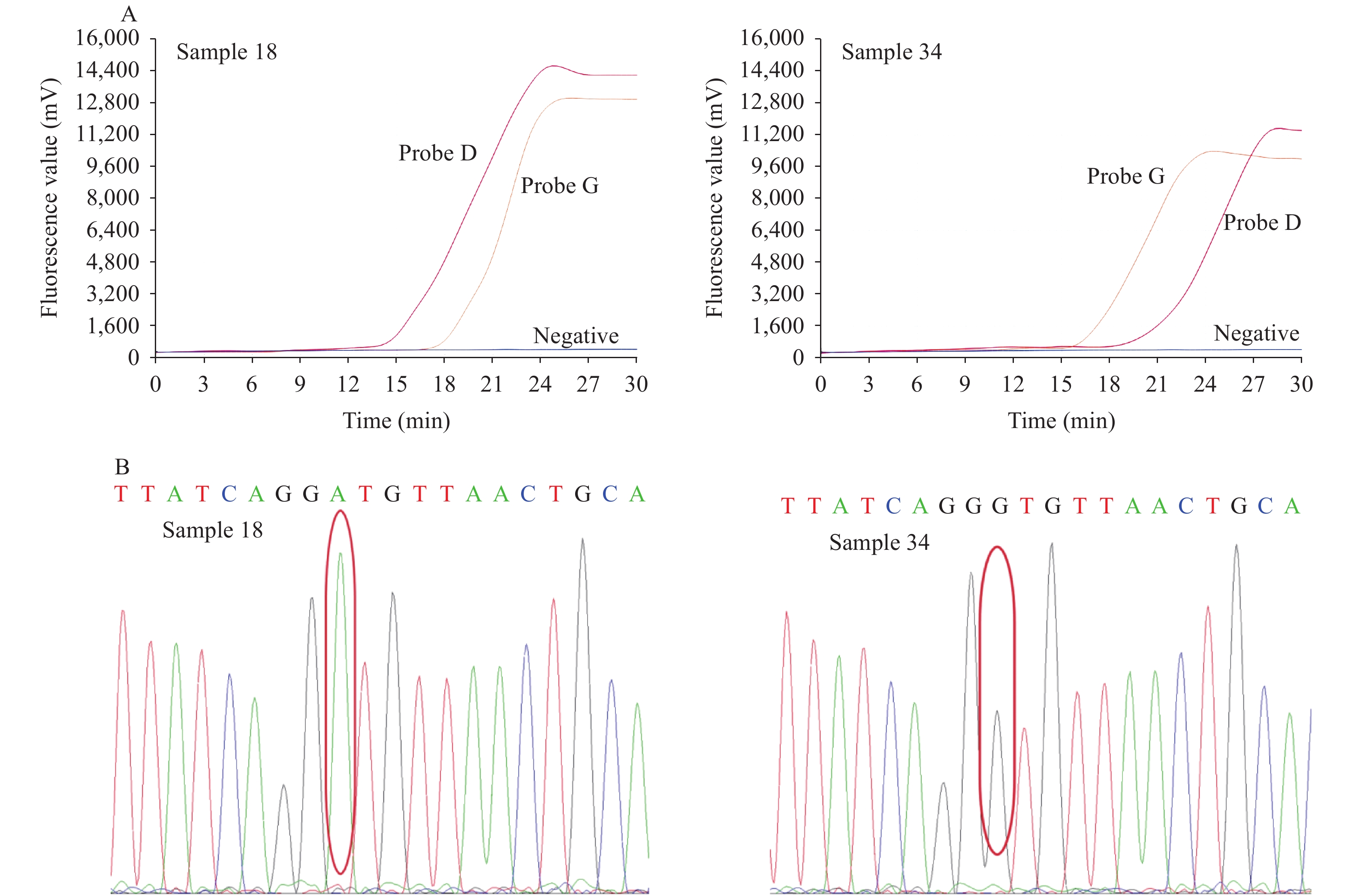 Figure 4.
Figure 4.Comparison of the PDRA assay and direct sequencing results. (A) The PDRA results of sample 18 and sample 34; (B) The direct sequencing results of sample 18 and sample 34.
Note: Sample 18 was S-D614 mutant, and sample 34 was S-G614 mutant. Sample 18 was S-D614 mutant, and sample 34 was S-G614 mutant.Type Sequencing PDRA Coincidence rate (%) S-D614 22 22 100 S-G614 31 31 100 Negative 10 10 100 Abbreviation: PDRA=probe-directed recombinase amplification. Table 2. Clinical sample typing results.
-
At present, COVID-19 is still spreading rapidly all over the world. The virus spike protein is one of the best-targeted molecules for the development of vaccines or monoclonal antibodies against this virus (15–16). Therefore, the mutation of S protein has special clinical significance. As one of the key mutations of S protein, the D614G mutation has been concerned by many researchers. D614G mutation was first found in Germany (17). After that, Becerra-Flores et al. suggested that S-G614 mutants are more pathogenic and have higher mortality (18). These characteristics make S-G614 mutants rapidly become the dominant species and expand around the world (3).
Few studies reported D614G detection. The D614G detection method designed by Hashemi et al. (14), actually detected the mutation to V615V (19). Whole-genome sequencing is a powerful but costly tool to identify D614G mutants, which cannot be applied in primary laboratories for sequencing a large number of samples. Considering the importance of D614G compared with other mutations, a simple and rapid method to detect this mutation is favorable. Prior to this, PDRA has been successfully used to detect heart disease-related SNP and prostate cancer-related SNP (12–13).
The two PDRA reactions were carried out separately for two reasons: 1) in order to achieve good sensitivity for two different mutants; 2) in order to make the results of PDRA easier to interpret. Through the experiment, the detection limit of the PDRA was found to be in the range of 103–107 copies/reaction. PDRA assay was not able to specifically distinguish the genotypes of the sample in the higher sample concentration (>107 copies) or led to false-negative results in the lower sample concentration (<103 copies). Adding two reaction systems in one tube was attempted for saving time and cost but failed in the end. A possible reason is that there might be a competition mechanism between the two probes that makes them unable to react.
Although the PDRA method has some limitations, compared with direct sequencing, PCR and LAMP, PDRA possess the advantages of simple experimental conditions, short turnaround time and appropriate sensitivity and specificity. PDRA assay needs only 30 minutes to complete the detection and requires no sophisticated equipment. The detection process of PDRA is also simple and easy to learn. In view of the characteristics of PDRA and further improvement, PDRA will facilitate the detection of D614G mutations in resource-limited settings, particularly in locations where the contamination of the vaccine strain (D614) needs to be monitored and differentiated with non-local circulating strains (G614) in China.
Conflicts of interest: No conflicts of interest were reported.
Availability of data and materials: The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
HTML
Clinical Sample Collection
Direct Sequencing of Clinical Samples
Design of Primer & Probes for the PDRA Assay
PDRA Assay
Interpretation of the PDRA Results
Analytical Sensitivity, Specificity, and Reproducibility of PDRA Assay
Detection of Clinical Samples by PDRA Assay
| Citation: |




 Download:
Download:




