-
Introduction: Influenza surveillance is necessary for detection of emerging variants of influenza viruses and determining their epidemiological and clinical significance. Vaccination and antiviral drugs remain the most useful ways to protect against seasonal influenza and its potentially severe consequences. This study describes the epidemiology, antigenic and genetic characteristics, and antiviral susceptibilities of influenza viruses isolated from the mainland of China during the April 1, 2019 through October 4, 2020.
Methods: All viruses analyzed were isolated and submitted by Chinese National Influenza Surveillance Network laboratories. The Chinese National Influenza Center performed antigenic analysis, sequencing, and antiviral resistance testing after propagation of the viruses. Antigenic characterizations were determined by hemagglutinin inhibition assay; next-generation sequencing was used for genetic analyses; phenotypic assay and next-generation sequencing were used for determining antiviral resistance.
Results: The influenza positivity rate declined significantly starting in late January 2020 and has remained low. There was no summer influenza peak season in south China. Influenza A(H3N2) and B/Victoria lineage viruses were the dominant subtype/lineage during April 1, 2019 through October 4, 2020. The majority of influenza viruses were antigenically and genetically similar to reference viruses representing components of vaccines for the 2020–2021 northern hemisphere influenza season. Nearly all seasonal influenza viruses were susceptible to oseltamivir and zanamivir.
Conclusions and Implications for Public Health Practice: Since the outbreak of COVID-19, the influenza positivity rate declined with implementation of strong COVID-19 control measures. The majority of circulating viruses are well matched with the current 2020–2021 northern hemisphere influenza vaccine viruses. Circulating seasonal influenza viruses were sensitive to neuraminidase inhibitors and Baloxavir marboxil. This study provided evidence for World Health Organization (WHO) recommendations for vaccine viruses, for prevention and control of influenza, and for clinical use of antiviral medications.
-
Influenza viruses evolve rapidly and escape natural or vaccine-induced immune responses by accumulating mutations within the surface glycoprotein genes for hemagglutinin (HA) and neuraminidase (NA) (1). The emergence of coronavirus disease 2019 (COVID-19) has had a huge impact on the world by shifting public health challenges and changing behaviors that even affect influenza activity.
Between April 2019 and September 2020, influenza activity was reported in all global regions. However, since April 2020, low levels of influenza activity have been reported — including from countries in the southern hemisphere temperate zone. Although influenza A(H1N1)pdm09, A(H3N2), and influenza B viruses co-circulated, the predominant circulating viruses varied by country and region. Overall, influenza A viruses were detected more often than influenza B viruses. Globally, co-circulation of A(H1N1)pdm09 and A(H3N2) viruses was evident in most countries, areas, and territories, but influenza A(H1N1)pdm09 viruses circulated in higher proportion than A(H3N2) viruses beginning in mid-January 2020. The B/Victoria lineage circulated in higher proportion than the B/Yamagata lineage viruses worldwide (2-3).
Each year, the WHO recommends compositions for the northern hemisphere influenza vaccine in February and for the southern hemisphere in September. Vaccination remains the best way to protect against seasonal influenza and its potentially severe consequences.
Influenza A(H3N2) and B/Victoria lineage viruses were the dominant subtype/lineage in the mainland of China during 2019–2020. To understand the variation of circulating seasonal influenza viruses and their match with vaccine virus strains, we analyzed the antigenic and genetic characteristics and antiviral susceptibilities of influenza viruses isolated from the mainland of China.
-
The Chinese National Influenza Surveillance Network includes 410 laboratories and 554 sentinel hospitals. The influenza surveillance year typically starts on week 14, which is around April 1, and lasts for an entire year. Sentinel hospitals report influenza-like illness (ILI) cases to the Chinese National Influenza Surveillance Information System (CNISIS) and collect respiratory specimens. Network laboratories test the specimens with real-time reverse transcriptase polymerase chain reaction (RT-PCR). Influenza positive specimens are propagated in Madin-Darby canine kidney (MDCK) cells and/or embryonated chicken eggs. Viruses are submitted to the Chinese National Influenza Center (CNIC) for further characterization.
Antigenic characterizations were assessed with HA inhibition (HI) assays, and genetic characterization was performed with next-generation sequencing. Influenza virus testing for antiviral resistance was conducted at CNIC using next-generation sequencing analysis, phenotypic assays, or with both tests (4). The viruses evaluated were isolated from specimens collected between week 14 in 2019 (April 1, 2019) and week 40 in 2020 (October 4, 2020).
-
For virus surveillance during April 1, 2019 through October 4, 2020 (surveillance week 14 in 2019 to week 40 in 2020), the percentage of specimens testing positive for influenza each week ranged from 0% to 46.5% in southern China and ranged from 0% to 47.3% in northern China. The positivity rate of specimens collected from ILI cases increased starting in week 40 in 2019 reaching a peak (46.5% for southern China and 47.3% for northern China) during the first week of 2020 and decreasing substantially after that. During week 8, the positivity rate in southern China decreased to 2.0%, and by week 10 in northern China, the influenza-positive rate declined to 1.3%. The positivity rate since then has been lower than that of the same period in previous years (5) and has remained low ever since (Figures 1 and 2).
 Figure 1.
Figure 1.Influenza positive tests reported by network laboratories in southern China since 2010 to 2020.
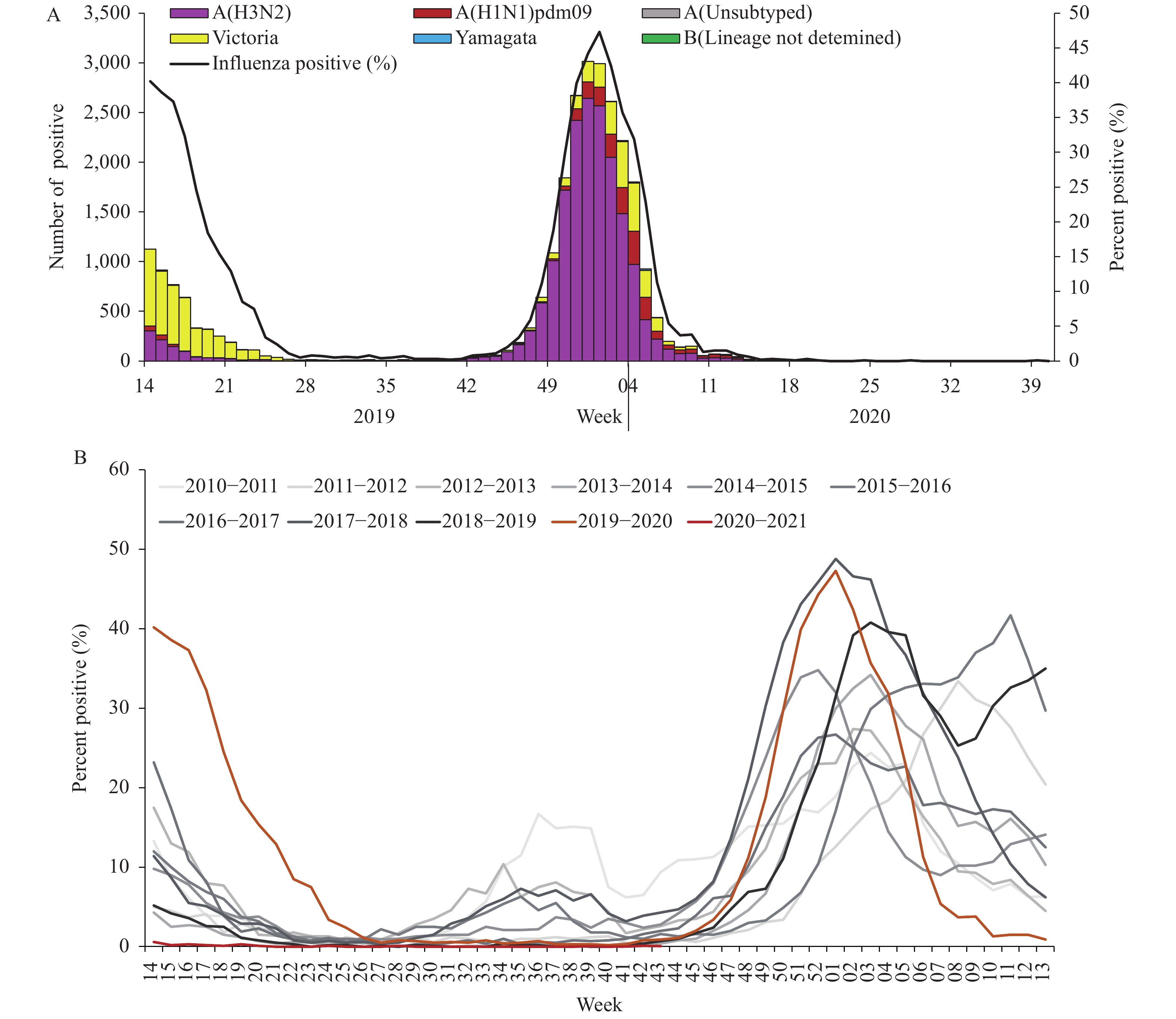 Figure 2.
Figure 2.Influenza positive tests reported by network laboratories in northern China since 2010 to 2020.
During the study period, network laboratories in southern China tested 423,466 specimens for influenza; among these specimens, 49,147 (11.6%) tested positive including 23,234 (47.3%) for influenza A and 25,913 (52.7%) for influenza B. Most of the positive samples were collected before March 2020. Among the 23,208 seasonal influenza A positive specimens that were subtyped, 2,830 (12.2%) were influenza A(H1N1)pdm09 and 20,378 (87.8%) were influenza A(H3N2). Among the 25,627 influenza B viruses for which lineage was determined, 25,477 (99.4%) belonged to the B/Victoria lineage and 150 (0.6%) belonged to the B/Yamagata lineage (Figure 1).
Network laboratories in northern China tested 216,874 specimens between April 1, 2019 and October 4, 2020. Among these, 26,759 (12.3%) were positive for influenza viruses — influenza A and influenza B viruses were 20,203 (75.5%) and 6,556 (24.5%), respectively, of the tested viruses. Among the 20,199 seasonal influenza A viruses that were subtyped, 2,091 (10.4%) were influenza A(H1N1)pdm09 and 18,108 (89.6%) were influenza A(H3N2). Influenza B lineage information was available for 6,547 influenza B viruses; 6,460 (98.7%) were B/Victoria lineage and 87 (1.3%) were B/Yamagata lineage (Figure 2).
CNIC tested the antigenic and genetic characteristics of influenza viruses between April 1, 2019 and October 4, 2020. A total of 653 A(H1N1)pdm09 viruses were analyzed with HI tests, and 521 viruses were antigenically analyzed with A/Guangdong-Maonan/SWL1536/2019, and 98.3% (512/521) were well inhibited by ferret antisera raised against A/Guangdong-Maonan /SWL1536/2019, the egg-propagated reference virus representing the A(H1N1)pdm09 component for the upcoming 2020–2021 winter season’s northern hemisphere influenza vaccination (2). Among the 205 viruses sequenced, phylogenetic analysis of HA gene segments determined that 199 (97.1%) belonged to genetic clade 6B.1A (Figure 3); 161 (78.5%) belonged to subclade 6B.1A5A, which evolved from clade 6B.1A. Subclade 6B.1A5A HA genes fall into 3 genetic groups: a progenitor 6B.1A5A subclade (22.9%) and 2 recently designated groups, 5A-187A (46.3%), with additional amino acid substitutions D187A and Q189E, and 5A-156K (9.3%), with an additional amino acid substitution N156K.
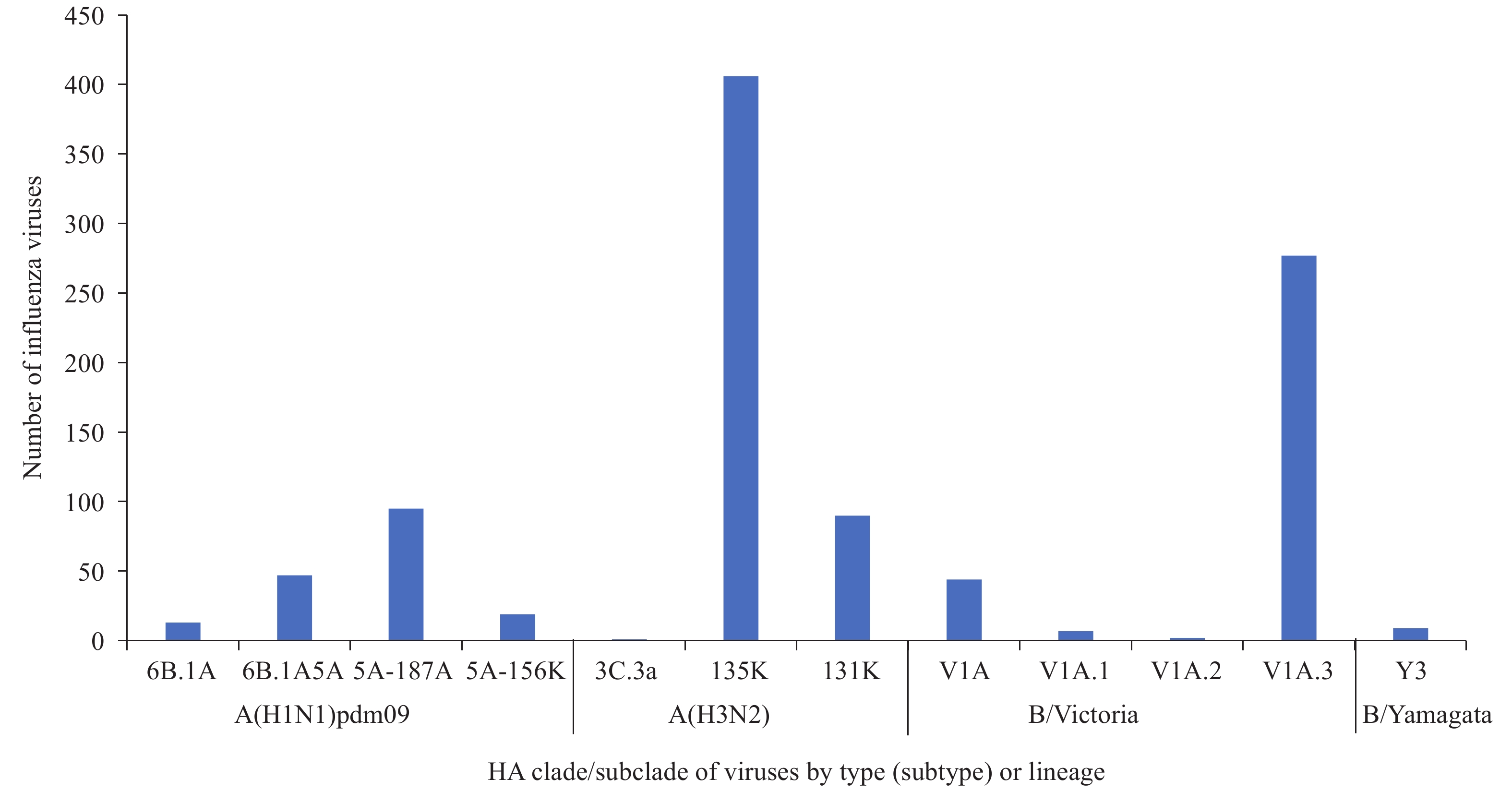 Figure 3.
Figure 3.Genetic characterization of influenza viruses in the mainland of China during April 1, 2019–October 4, 2020.
Antigenic characterization of 1,410 A(H3N2) viruses were conducted using HI tests that used guinea pig red blood cells (RBCs) in the presence of oseltamivir, and 63 viruses underwent antigenic analysis with A/Hong Kong/2671/2019 where the results showed that 79.4% (50/63) of virus isolates were well inhibited by ferret antisera raised against A/Hong Kong/2671/2019, the egg-propagated reference virus representing the A(H3N2) component for the 2020–2021 northern hemisphere influenza vaccine — a higher proportion than the last influenza season (6). Among the 502 viruses sequenced, phylogenetic analysis of the HA gene segments determined that 501 (99.8%) viruses belonged to genetic clade 3C.2a, and only 1 virus belonged to clade 3C.3a. Multiple subclades within the 3C.2a clade co-circulated with viruses belonging to subclade 3C.2a1b, the majority subclade. Subclade 3C.2a1b viruses includes viruses having either T135K or T131K amino acid substitutions in their HA protein; 406 (80.9%) belonged to 3C.2a1b+T135K and 90 (17.9%) belonged to 3C.2a1b+T131K (Figure 3).
A total of 2,070 B/Victoria lineage viruses were antigenically analyzed with the HI test. Overall, 1,012 viruses underwent antigenic analysis with B/Washington /02/2019, and 92.4% (935/1012) were similar to B/Washington/02/2019, the egg-propagated reference virus representing the B/Victoria lineage component for the 2020–2021 northern hemisphere influenza vaccine. Significant genetic diversity was seen in B/Victoria lineage cocirculating viruses. In the HA proteins, viruses with a 2 amino acid deletions (positions 162 and 163) belonged to subclade V1A.1, and viruses with a 3 amino acid deletions (positions 162–164) belonged to subclade V1A.2; subclade V1A.3 shared a triple amino acid deletion and had an additional substitution at K136E (2,7). Among the 330 virus HA gene segments sequenced and phylogenetically analyzed, 44 (13.3%) belonged to genetic clade V1A, 7 (2.1%) belonged to subclade V1A.1, 2 (0.6%) belonged to subclade V1A.2, and 277 (83.9%) belonged to subclade V1A.3 (Figure 3).
Few B/Yamagata lineage viruses were detected during the study period. Antigenic characterization of the 12 B/Yamagata lineage viruses by HI test indicated that 11 (91.7%) viruses were similar to the egg-propagated B/Phuket/3073/2013, the reference virus representing the B/Yamagata lineage component of quadrivalent vaccines for the northern hemisphere influenza season. Phylogenetic analysis of 9 (100%) influenza B/Yamagata lineage viruses determined that the HA gene segments belonged to clade Y3 (Figure 3).
CNIC then tested 41,66 influenza viruses collected in the mainland of China for resistance to oseltamivir and zanamivir, including 664 influenza A(H1N1)pdm09, 1,419 influenza A(H3N2), 2,071 influenza B/Victoria, and 12 influenza B/Yamagata viruses. Overall, 3 influenza A(H1N1)pdm09 viruses showed highly reduced inhibition by oseltamivir; 2 had a H275Y amino acid substitution and 1 had a H275H/Y mixed substitution in the NA protein. In addition, 1 influenza B/Victoria lineage virus had a G243D amino acid substitution and exhibited reduced inhibition by oseltamivir and highly reduced inhibition by zanamivir; 1 influenza B/Victoria lineage virus showed reduced susceptibility to zanamivir with substitution D198N in the NA protein. All sequenced seasonal influenza viruses do not carry any reported resistant mutations to Baloxavir marboxil. All sequenced influenza A(H1N1)pdm09 and influenza A(H3N2) viruses were resistant to adamantanes, which was consistent with the current recommendation to avoid use of these drugs against influenza.
-
Unlike previous years, influenza activity began to decline sharply after the epidemic peaked in the first week of 2020. Lower levels of influenza activity have also been reported globally during the April to September period in 2020 compared with recent years (3). This decrease in activity could be due to the COVID-19 pandemic response, as the government implemented a series of prevention and control measures, including home quarantine and wearing masks, to stop spread of COVID-19.
In late February 2020, the WHO issued its recommendations for the 2020–2021 northern hemisphere influenza vaccines. Egg-based influenza trivalent vaccines will use an A/Guangdong-Maonan/SWL1536/2019(H1N1)pdm09-like virus, an A/Hong Kong/2671/2019 (H3N2)-like virus, and a B/Washington/02/2019-like virus (B/Victoria lineage). For quadrivalent vaccines, an additional component, the B/Phuket/3073/2013-like virus (B/Yamagata lineage), was recommended (2). Except for the B/Yamagata lineage, all vaccine components were updated from the 2019–2020 northern hemisphere influenza vaccine formulation.
Most of these viruses belonged to subclade 6B.1A5A and contained amino acid substitutions D187A and Q189E (e.g. A/Guangdong-Maonan/SWL1536/2019) in their HA proteins. Although antigenic analyses do not detect all antigenic differences in these A(H1N1)pdm09 viruses, amino acid substitutions acquired in the circulating viruses were within the antigenic HA epitopes (8). Almost all influenza A(H1N1)pdm09 viruses were well inhibited by ferret antisera raised against A/Guangdong-Maonan/SWL1536/2019, but group 5A-156K viruses reacted poorly with these antisera and were antigenically distinct, and the 5A-156K group has increased rapidly in many countries since December 2019 (3). The majority of circulating A(H3N2) viruses belonged to subclade 3C.2a1b. Among A(H3N2) viruses circulating globally, there were regional differences in the proportions of subclades circulating, and viruses with 2a1b+T135K subclade (e.g. A/Hong Kong/2671/2019) are the most prevalent in the mainland of China. Because most of the B/Victoria lineage viruses belonged to subclade V1A.3 (e.g. B/ Washington/02/2019), they were well inhibited by post-infection ferret antisera raised against B/Washington/02/2019 viruses. Most B/Yamagata lineage viruses were antigenically and genetically similar to the reference viruses representing the components of vaccines for the 2019−2021 northern hemisphere influenza season. It is worth noting that vaccine effectiveness cannot be determined solely based on similarity between circulating viruses and vaccine reference viruses because there are many other factors influencing vaccine effectiveness.
Based on our analysis, >99% of seasonal influenza viruses were susceptible to oseltamivir and zanamivir. Only 3 influenza A(H1N1)pdm09 viruses were resistant and included the H275Y or H275H/Y amino acid substitution in the NA protein that has previously been associated with highly reduced susceptibility to oseltamivir. The H275Y amino acid substitution in A(H1N1)pdm09 viruses is considered clinically relevant and leads to reduced treatment efficacy (4). An influenza B/Victoria lineage virus showed reduced inhibition by oseltamivir and highly reduced inhibition by zanamivir; this virus had a G243D amino acid substitution in the NA protein, a substitution that has not been previously described. The G243S/G amino acid substitution, which was reported in B/Victoria-lineage viruses, is associated with reduced susceptibility to zanamivir (9). We should conduct additional evaluation to clarify the significance of this substitution. Anti-influenza drugs are effective as post-exposure prophylaxis and treatments for influenza virus infection. It is important to continuously monitor antiviral resistance of circulating influenza viruses.
The COVID-19 pandemic has changed lifestyles, improved public health preventative measures, and had an impact on other respiratory infectious diseases including influenza. Surveillance and research on influenza should be strengthened, including human infection with zoonotic influenza, to better predict and prepare for the next pandemic.
HTML
| Citation: |




 Download:
Download:




